
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,448,456 – 14,448,842 |
| Length | 386 |
| Max. P | 0.999788 |

| Location | 14,448,456 – 14,448,576 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -32.74 |
| Energy contribution | -34.10 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14448456 120 - 27905053 AUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCCACCAAAUUGACGAAGUUCCCCUUCAUGAGUGCCCCGGGGUGACGGCAUUCAGCCAAUUGCACAGACAAUAACCAGGAAUA ..(((((((...((((.(((((((.....)))))))........(((((.(((((.....)))).((((((((....(....)))))))))).))))))))).........))))))).. ( -36.20) >DroSec_CAF1 5710 120 - 1 AUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCCACCAAAUUGGCGGAGUUCCCCUUCAUGAGUGCCCCAGGGUGACGGCAUUCAGCCAAUUGCGCAGACAAUAACCAGGAAUG ..(((((((...((((.(((((((.....)))))))........(((((((((((.....)))).((((((((....(....)))))))))))))))))))).........))))))).. ( -42.40) >DroSim_CAF1 6267 120 - 1 AUUUUUUGGCACGUGCAGUGUCCACUUUCUGGGCACACCCACCAAAUUGGCGGAGUUCCCCUUCAUGAGUGCCCCAGGGUGACGGCAUUCAGCCAAUUGCGCAGACAAUAACCAGGAAUG ..(((((((...((((.(((((((.....)))))))........(((((((((((.....)))).((((((((....(....)))))))))))))))))))).........))))))).. ( -41.80) >DroEre_CAF1 5551 97 - 1 AUUUUUCGGCACGUGCAGUGUCCACUUUCUGGACACACCCACAAAAUUGGAGGAGUUCCCCUUCAUGAGUGCCCUGGGGUGACGGCAUUCAGCCAAU----------------------- .......(((..(((..(((((((.....)))))))...))).....((((((......))))))((((((((....(....))))))))))))...----------------------- ( -37.30) >DroYak_CAF1 6343 119 - 1 AUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCCACAAAAUUGGCGGAGUUUGCCUUCAUGACUGCCCAGGGG-GACUGCAUUCAGCCAAUUGCGCAGACAAUAACCACGAAUG (((.((((((..(((..(((((((.....)))))))...)))..(((((((((((.....)))).(((.(((...((..-..))))).)))))))))))).)))).)))........... ( -36.30) >consensus AUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCCACCAAAUUGGCGGAGUUCCCCUUCAUGAGUGCCCCGGGGUGACGGCAUUCAGCCAAUUGCGCAGACAAUAACCAGGAAUG ............((((.(((((((.....)))))))........(((((((((((.....)))).((((((((....(....)))))))))))))))))))).................. (-32.74 = -34.10 + 1.36)


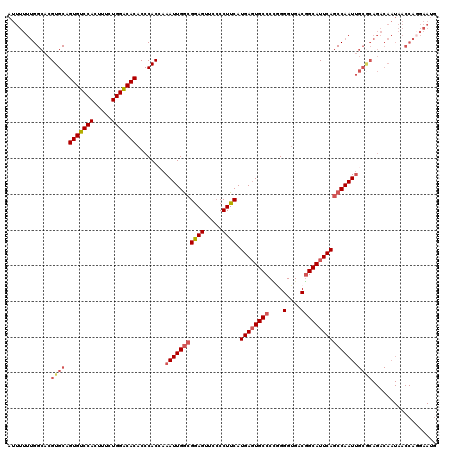
| Location | 14,448,496 – 14,448,613 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.81 |
| Mean single sequence MFE | -41.86 |
| Consensus MFE | -33.36 |
| Energy contribution | -34.40 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14448496 117 + 27905053 ACCCCGGGGCACUCAUGAAGGGGAACUUCGUCAAUUUGGUGGGUGUGUCCAGAAAGUGGACACUGCACGUGCCAAAAAAUGGUUUAGAAAGUGCCCAAAGGCACUUUGUGGCAAAA---A ...(((.(((((..((((((.....)))))).......(((.(.(((((((.....)))))))).))))))))......))).(((.((((((((....)))))))).))).....---. ( -41.80) >DroSec_CAF1 5750 117 + 1 ACCCUGGGGCACUCAUGAAGGGGAACUCCGCCAAUUUGGUGGGUGUGUCCAGAAAGUGGACACUGCACGUGCCAAAAAAUUGUUUAGAAAGUGCCCAAAGGCACUUUGUGGCACAA---A ((((.((((..(((......)))..))))(((.....)))))))(((((((.....))))))).....((((((.............((((((((....)))))))).))))))..---. ( -44.44) >DroSim_CAF1 6307 117 + 1 ACCCUGGGGCACUCAUGAAGGGGAACUCCGCCAAUUUGGUGGGUGUGCCCAGAAAGUGGACACUGCACGUGCCAAAAAAUGGUUUAGAAAGUGCCCAAAGGCACUUUGUGGCACAA---A ...(((((((((((((.((((((....)).))...)).)))))))).)))))...(((.......)))((((((.............((((((((....)))))))).))))))..---. ( -44.94) >DroEre_CAF1 5568 117 + 1 ACCCCAGGGCACUCAUGAAGGGGAACUCCUCCAAUUUUGUGGGUGUGUCCAGAAAGUGGACACUGCACGUGCCGAAAAAUGGUUUAGAAAGUGGCCAAAGGCACUUUGUGGCACAA---A ....(((.(((((((..((((((....))))....))..)))))))(((((.....))))).)))...((((((((......)))(.((((((.(....).)))))).))))))..---. ( -38.40) >DroYak_CAF1 6383 119 + 1 -CCCCUGGGCAGUCAUGAAGGCAAACUCCGCCAAUUUUGUGGGUGUGUCCAGAAAGUGGACACUGCACGUGCCAAAAAAUGGUUUAGAAAGUGCCCAAAGGCACUUCGUGGCAAAAAAAA -((....))..((((((((((((.((.((((.......))))))(((((((.....)))))))......)))).................(((((....)))))))))))))........ ( -39.70) >consensus ACCCCGGGGCACUCAUGAAGGGGAACUCCGCCAAUUUGGUGGGUGUGUCCAGAAAGUGGACACUGCACGUGCCAAAAAAUGGUUUAGAAAGUGCCCAAAGGCACUUUGUGGCACAA___A .((((....((....))..))))......((((.(((((((.(((((((((.....)))))...)))).)))))))...........((((((((....)))))))).))))........ (-33.36 = -34.40 + 1.04)



| Location | 14,448,496 – 14,448,613 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.81 |
| Mean single sequence MFE | -38.21 |
| Consensus MFE | -34.66 |
| Energy contribution | -34.58 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
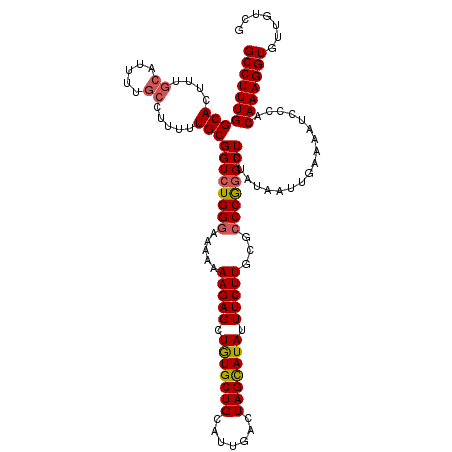
>3R_DroMel_CAF1 14448496 117 - 27905053 U---UUUUGCCACAAAGUGCCUUUGGGCACUUUCUAAACCAUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCCACCAAAUUGACGAAGUUCCCCUUCAUGAGUGCCCCGGGGU .---....(((..((((((((....))))))))..............((((((((..(((((((.....)))))))...))).........((((.....))))....)))))....))) ( -37.50) >DroSec_CAF1 5750 117 - 1 U---UUGUGCCACAAAGUGCCUUUGGGCACUUUCUAAACAAUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCCACCAAAUUGGCGGAGUUCCCCUUCAUGAGUGCCCCAGGGU .---..((((((.((((((((....)))))))).............)))))).....(((((((.....)))))))((((.......(((.((..(((........)))..))))))))) ( -39.25) >DroSim_CAF1 6307 117 - 1 U---UUGUGCCACAAAGUGCCUUUGGGCACUUUCUAAACCAUUUUUUGGCACGUGCAGUGUCCACUUUCUGGGCACACCCACCAAAUUGGCGGAGUUCCCCUUCAUGAGUGCCCCAGGGU (---(((.....))))..(((((.((((((((......(((...((((((....)).(((((((.....)))))))......)))).))).((((.....))))..)))))))).))))) ( -38.80) >DroEre_CAF1 5568 117 - 1 U---UUGUGCCACAAAGUGCCUUUGGCCACUUUCUAAACCAUUUUUCGGCACGUGCAGUGUCCACUUUCUGGACACACCCACAAAAUUGGAGGAGUUCCCCUUCAUGAGUGCCCUGGGGU .---....(((..((((((((...)).)))))).....(((......((((((((..(((((((.....)))))))...))).....((((((......))))))...))))).)))))) ( -37.60) >DroYak_CAF1 6383 119 - 1 UUUUUUUUGCCACGAAGUGCCUUUGGGCACUUUCUAAACCAUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCCACAAAAUUGGCGGAGUUUGCCUUCAUGACUGCCCAGGGG- .......(((.((((((((((....)))))))......(((.....)))..))))))(((((((.....))))))).(((.......(((.(.((((.........)))).))))))).- ( -37.91) >consensus U___UUGUGCCACAAAGUGCCUUUGGGCACUUUCUAAACCAUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCCACCAAAUUGGCGGAGUUCCCCUUCAUGAGUGCCCCGGGGU .........((..((((((((....))))))))..............((((((((..(((((((.....)))))))...))).........((((.....))))....)))))...)).. (-34.66 = -34.58 + -0.08)



| Location | 14,448,536 – 14,448,642 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.77 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -25.90 |
| Energy contribution | -26.46 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14448536 106 + 27905053 GGGUGUGUCCAGAAAGUGGACACUGCACGUGCCAAAAAAUGGUUUAGAAAGUGCCCAAAGGCACUUUGUGGCAAAA---AACAUUAUGUUAUAGACACGG-----------UAUGUCAGU ....(((((((.....)))))))((.(((((((..............((((((((....))))))))(((......---(((.....))).....)))))-----------))))))).. ( -33.90) >DroSec_CAF1 5790 113 + 1 GGGUGUGUCCAGAAAGUGGACACUGCACGUGCCAAAAAAUUGUUUAGAAAGUGCCCAAAGGCACUUUGUGGCACAA---AACAUUAUGUUAUGGACAUGGGC----ACAUGGAUGUCAGU ..(((((((((.....))))))).))..(((((......(((((((.((((((((....)))))))).))).))))---.....((((((...)))))))))----))............ ( -38.30) >DroSim_CAF1 6347 113 + 1 GGGUGUGCCCAGAAAGUGGACACUGCACGUGCCAAAAAAUGGUUUAGAAAGUGCCCAAAGGCACUUUGUGGCACAA---AACAUUAUGUUAUGGACAUGGGC----ACAUGGAUGUCGGU ..(((((((((....(((.......)))((((((.............((((((((....)))))))).))))))..---..................)))))----)))).......... ( -37.44) >DroEre_CAF1 5608 117 + 1 GGGUGUGUCCAGAAAGUGGACACUGCACGUGCCGAAAAAUGGUUUAGAAAGUGGCCAAAGGCACUUUGUGGCACAA---AAAGUGAUGUUAUGGACACAGGAGCGAGCAUGGAUGCCAGU .((((((((((.....)))))))....(((((((.....((((((..((((((.(....).))))))((((((((.---....)).)))))))))).))....)).)))))...)))... ( -34.80) >DroYak_CAF1 6422 116 + 1 GGGUGUGUCCAGAAAGUGGACACUGCACGUGCCAAAAAAUGGUUUAGAAAGUGCCCAAAGGCACUUCGUGGCAAAAAAAAAAAUUAUGUUAUGGACAUAGGC----ACAUGGAUGUCAGU ..(((((((((.....))))))).))..(((((.................(((((....)))))(((((((((.((.......)).)))))))))....)))----))............ ( -34.20) >consensus GGGUGUGUCCAGAAAGUGGACACUGCACGUGCCAAAAAAUGGUUUAGAAAGUGCCCAAAGGCACUUUGUGGCACAA___AACAUUAUGUUAUGGACAUGGGC____ACAUGGAUGUCAGU ...((((((((....(((.......)))((((((.............((((((((....)))))))).)))))).................))))))))..................... (-25.90 = -26.46 + 0.56)

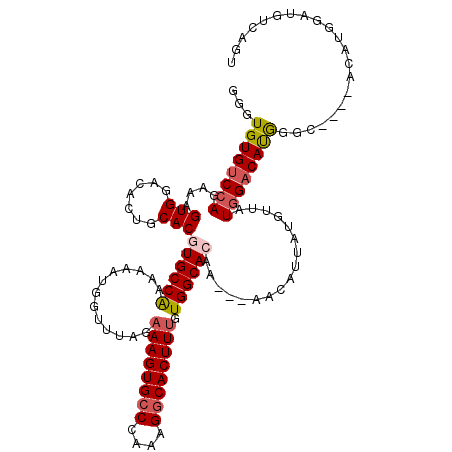

| Location | 14,448,536 – 14,448,642 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.77 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -28.06 |
| Energy contribution | -28.14 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14448536 106 - 27905053 ACUGACAUA-----------CCGUGUCUAUAACAUAAUGUU---UUUUGCCACAAAGUGCCUUUGGGCACUUUCUAAACCAUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCC ...(((((.-----------..)))))..............---...(((.((((((((((....)))))))).....(((.....)))...)))))(((((((.....))))))).... ( -30.60) >DroSec_CAF1 5790 113 - 1 ACUGACAUCCAUGU----GCCCAUGUCCAUAACAUAAUGUU---UUGUGCCACAAAGUGCCUUUGGGCACUUUCUAAACAAUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCC .........(((((----(((.((((.....))))..((((---(.(((((.(((((...)))))))))).....))))).......))))))))..(((((((.....))))))).... ( -38.30) >DroSim_CAF1 6347 113 - 1 ACCGACAUCCAUGU----GCCCAUGUCCAUAACAUAAUGUU---UUGUGCCACAAAGUGCCUUUGGGCACUUUCUAAACCAUUUUUUGGCACGUGCAGUGUCCACUUUCUGGGCACACCC .........(((((----(((...(..((.(((.....)))---.))..)...((((((((....))))))))..............))))))))..(((((((.....))))))).... ( -36.60) >DroEre_CAF1 5608 117 - 1 ACUGGCAUCCAUGCUCGCUCCUGUGUCCAUAACAUCACUUU---UUGUGCCACAAAGUGCCUUUGGCCACUUUCUAAACCAUUUUUCGGCACGUGCAGUGUCCACUUUCUGGACACACCC ....((((..(((..(((....)))..)))...........---..(((((..((((((..(((((.......))))).))))))..))))))))).(((((((.....))))))).... ( -30.90) >DroYak_CAF1 6422 116 - 1 ACUGACAUCCAUGU----GCCUAUGUCCAUAACAUAAUUUUUUUUUUUGCCACGAAGUGCCUUUGGGCACUUUCUAAACCAUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCC .........(((((----((((((((.....))))).................((((((((....))))))))..............))))))))..(((((((.....))))))).... ( -37.10) >consensus ACUGACAUCCAUGU____GCCCAUGUCCAUAACAUAAUGUU___UUGUGCCACAAAGUGCCUUUGGGCACUUUCUAAACCAUUUUUUGGCACGUGCAGUGUCCACUUUCUGGACACACCC ...(((((..............)))))..................(((((((.((((((((....)))))))).............)))))))....(((((((.....))))))).... (-28.06 = -28.14 + 0.08)

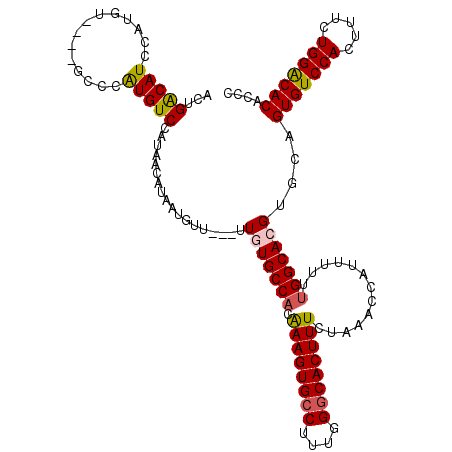

| Location | 14,448,576 – 14,448,682 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.79 |
| Mean single sequence MFE | -16.83 |
| Consensus MFE | -12.08 |
| Energy contribution | -11.72 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14448576 106 - 27905053 UCGACCAUUUCUAACUAAAAGCCCCCCCCCCCACACACACACUGACAUA-----------CCGUGUCUAUAACAUAAUGUU---UUUUGCCACAAAGUGCCUUUGGGCACUUUCUAAACC .................................................-----------..(((.(...(((.....)))---....).)))((((((((....))))))))....... ( -14.20) >DroSec_CAF1 5830 108 - 1 UCGACCAUUUCAAACUAAAAGCCCCC-----CCCACACACACUGACAUCCAUGU----GCCCAUGUCCAUAACAUAAUGUU---UUGUGCCACAAAGUGCCUUUGGGCACUUUCUAAACA ..........................-----......((((..(((((..((((----.............)))).)))))---.))))....((((((((....))))))))....... ( -19.02) >DroSim_CAF1 6387 108 - 1 UCGACCAUUUCAAACUAAAAGCCCCC-----CACACACACACCGACAUCCAUGU----GCCCAUGUCCAUAACAUAAUGUU---UUGUGCCACAAAGUGCCUUUGGGCACUUUCUAAACC ..........................-----......((((..(((((..((((----.............)))).)))))---.))))....((((((((....))))))))....... ( -18.12) >DroEre_CAF1 5648 106 - 1 UCGACCAUUUCAAACUAAAAGCC-C----------CACACACUGGCAUCCAUGCUCGCUCCUGUGUCCAUAACAUCACUUU---UUGUGCCACAAAGUGCCUUUGGCCACUUUCUAAACC ....................(((-.----------....(((((((....(((..(((....)))..))).(((.......---.))))))....)))).....)))............. ( -15.40) >DroYak_CAF1 6462 106 - 1 UCGACCAUUUCAAACUAAAAGCCCC----------CACACACUGACAUCCAUGU----GCCUAUGUCCAUAACAUAAUUUUUUUUUUUGCCACGAAGUGCCUUUGGGCACUUUCUAAACC ..........((((..(((((....----------..((((.((.....)))))----)..(((((.....)))))..)))))..))))....((((((((....))))))))....... ( -17.40) >consensus UCGACCAUUUCAAACUAAAAGCCCCC_____C_CACACACACUGACAUCCAUGU____GCCCAUGUCCAUAACAUAAUGUU___UUGUGCCACAAAGUGCCUUUGGGCACUUUCUAAACC ...........................................(((((..............)))))..........................((((((((....))))))))....... (-12.08 = -11.72 + -0.36)


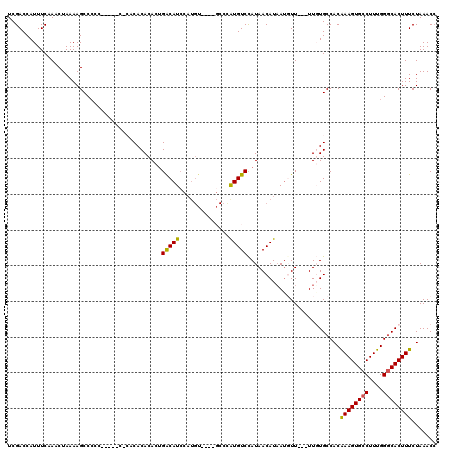
| Location | 14,448,642 – 14,448,762 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.96 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -20.68 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14448642 120 - 27905053 CCCGGGCUUUAUAAUUGAAAAUCCCACAAAGGUGUUGUCGUUGCUUUGUGACUAAAUUGAAUUUAGUGAUCGCUUUUUUAUCGACCAUUUCUAACUAAAAGCCCCCCCCCCCACACACAC ...(((((((......((((....(((....)))..((((.......((((((((((...))))))...))))........))))..))))......)))))))................ ( -21.96) >DroSec_CAF1 5903 115 - 1 CCCAGGCUUUAUAAUUGAAAAUCCCACAAAGGUGUUGUCGUUGCUUUGUGACUAAAUUGAAUUUAGUAAUCGCUUUUUUAUCGACCAUUUCAAACUAAAAGCCCCC-----CCCACACAC ....((((((....((((((....(((....)))..((((.......((((((((((...))))))...))))........))))..))))))....))))))...-----......... ( -20.56) >DroSim_CAF1 6460 115 - 1 CCCGGGCUUUAUAAUUGAAAAUCCCACAAAGGUGUUGUCGUUGCUUUGUGACUAAAUUGAAUUUAGUAAUCGCUUUUUUAUCGACCAUUUCAAACUAAAAGCCCCC-----CACACACAC ...(((((((....((((((....(((....)))..((((.......((((((((((...))))))...))))........))))..))))))....)))))))..-----......... ( -23.96) >DroEre_CAF1 5725 109 - 1 ACCGGGCUUUAUAAUUGGAAAUCCCACAAAGGUGUUGUCGUUGCUUUGUGACUAAAUUGAAUUUAGUAAUCGCUUUUUUAUCGACCAUUUCAAACUAAAAGCC-C----------CACAC ...(((((((....((((((....(((....)))..((((.......((((((((((...))))))...))))........))))..))))))....))))))-)----------..... ( -23.46) >DroYak_CAF1 6538 110 - 1 CCCGGGCUUUAUAAUUGGAAAUCCCACAAAGGUGUUUUCGUUGCUUUGUGACUAAAUUGAAUUUAGUAAUCGCUUUUUUAUCGACCAUUUCAAACUAAAAGCCCC----------CACAC ...(((((((....((((((....(((((((............)))))))(((((((...)))))))....................))))))....))))))).----------..... ( -23.30) >consensus CCCGGGCUUUAUAAUUGAAAAUCCCACAAAGGUGUUGUCGUUGCUUUGUGACUAAAUUGAAUUUAGUAAUCGCUUUUUUAUCGACCAUUUCAAACUAAAAGCCCCC_____C_CACACAC ...(((((((....((((((....(((((((............)))))))(((((((...)))))))....................))))))....)))))))................ (-20.68 = -21.04 + 0.36)



| Location | 14,448,722 – 14,448,842 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -32.20 |
| Energy contribution | -32.52 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14448722 120 - 27905053 GCCUUUGGCACUUUGCAUUUUGCCUUUUUGCGGUCUGGGAAAAAAAGAGCUGUGCUGCAUUGACUAGCAUAUUUCUUGCGCCCGGGCUUUAUAAUUGAAAAUCCCACAAAGGUGUUGUCG ((((((((((....((.....)).....)))((((((((.....(((((.(((((((.......))))))).)))))...))))))))..................)))))))....... ( -36.30) >DroSec_CAF1 5978 120 - 1 GCCUUUGGCACUUUGCAUUUUGCCUUUUUGCGGUCUGGGAAAAAAAGAGCUAUGCUGCAUUGACUAGCAUAUUUCUUGCACCCAGGCUUUAUAAUUGAAAAUCCCACAAAGGUGUUGUCG ((((((((((....((.....)).....)))((((((((.....(((((.(((((((.......))))))).)))))...))))))))..................)))))))....... ( -35.90) >DroSim_CAF1 6535 120 - 1 GCCUUUGGCACUUUGCAUUUUGCCUUUUUGCGGUCUGGGAAAAAAAGAGCUGUGCUGCAUUGACUAGCAUAUUUCUUGCGCCCGGGCUUUAUAAUUGAAAAUCCCACAAAGGUGUUGUCG ((((((((((....((.....)).....)))((((((((.....(((((.(((((((.......))))))).)))))...))))))))..................)))))))....... ( -36.30) >DroEre_CAF1 5794 118 - 1 GCCUUUGGCACUUUGCAUUUUGCCUUUUUGCGGUAUGGGAAA--AAGAGCUGUGCUGCAUUGACUAGCAUAUUUCUUUCGACCGGGCUUUAUAAUUGGAAAUCCCACAAAGGUGUUGUCG (((((((((.....))....((((.......))))(((((.(--(((((.(((((((.......))))))).))))))...((((.........))))...))))))))))))....... ( -31.80) >DroYak_CAF1 6608 114 - 1 GCCUUUGGCACUU------UUGCCUUUUUGCGGUCUGGGAAAAAAAGAGCUGUGCUGCAUUGACUAGUAUAUUUCUUUCGCCCGGGCUUUAUAAUUGGAAAUCCCACAAAGGUGUUUUCG ((....((((...------.)))).....))((((((((....((((((.(((((((.......))))))).))))))..))))))))........(((((..((.....))..))))). ( -34.30) >consensus GCCUUUGGCACUUUGCAUUUUGCCUUUUUGCGGUCUGGGAAAAAAAGAGCUGUGCUGCAUUGACUAGCAUAUUUCUUGCGCCCGGGCUUUAUAAUUGAAAAUCCCACAAAGGUGUUGUCG ((((((((((....((.....)).....)))((((((((.....(((((.(((((((.......))))))).)))))...))))))))..................)))))))....... (-32.20 = -32.52 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:38 2006