| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,446,376 – 14,446,486 |
| Length | 110 |
| Max. P | 0.979719 |

| Location | 14,446,376 – 14,446,486 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -20.88 |
| Energy contribution | -21.76 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14446376 110 + 27905053 UUUGUUUGGUCCAA----------AAGAAGAGAAUGUUUUUUGUUUUGAAAACAUUUUUAUUUACUUUUAUUGUUUGAUAACUUGAAAAGAUUGGGGGAAAACCGGACUAAGAAAGUGUU ....((((((((..----------.(((.((((((((((((......)))))))))))).))).(((((...(((....)))...)))))....((......))))))))))........ ( -25.40) >DroSec_CAF1 3502 108 + 1 UUAGUUUGGUCCAA----------AAGAAGAGAAUGUUUUUUGUUUUGAAAACAUUUUUAUUUACUUUUAUUGUUUGAUAACUUGAAAAGGUUGGGG--AAACCGGACCAAGAAAGUGUU ....((((((((((----------(((((((((((((((((......))))))))))))..)).))))).........((((((....))))))((.--...))))))))))........ ( -31.70) >DroSim_CAF1 3815 108 + 1 UUAGUUUCGUCCAA----------AAGAAGAGAAUGUUUUUUGUUUUGAAAACAUUUUUAUUUACUUUUAUUGUUUGAUAACUUGAAAAGGUUGGGG--AAACCGGACCAAGAAAGUGUU ..(((((((..(((----------.((((((((((((((((......)))))))))))......))))).)))..))).))))......((((.((.--...)).))))........... ( -27.80) >DroEre_CAF1 3462 117 + 1 UUAGUUUGGUCCAAACCGGUGACAAAGAAAAGAAUGCGUUUUGUUUUGAAAACACU-UUAGUUACUUUUAUUGUGUGAUAACUUGAAAAGGUUGGGG--AAACCGGACCAAGAAAGUAUU ....((((((((.....((((((((((....(....)(((((......))))).))-)).))))))............((((((....))))))((.--...))))))))))........ ( -29.50) >DroYak_CAF1 3648 118 + 1 UUAGUUUGGUCCAAAACGGUGACAAAGAGAAGAAUGCGUUUUGUUUUGGAAACAUUUUUAGUUACUUUUAUUGUAUGACAACUUUAAAAGGUUGGUG--AAACCGGACCAAGAAAUUGUU ....((((((((......(((((.((((.((((.....)))).))))(....).......)))))(((((..((......))..)))))((((....--.))))))))))))........ ( -25.90) >consensus UUAGUUUGGUCCAA__________AAGAAGAGAAUGUUUUUUGUUUUGAAAACAUUUUUAUUUACUUUUAUUGUUUGAUAACUUGAAAAGGUUGGGG__AAACCGGACCAAGAAAGUGUU ....((((((((.................((((((((((((......)))))))))))).....(((((...((......))...)))))...((.......))))))))))........ (-20.88 = -21.76 + 0.88)


| Location | 14,446,376 – 14,446,486 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -16.95 |
| Consensus MFE | -13.30 |
| Energy contribution | -13.74 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14446376 110 - 27905053 AACACUUUCUUAGUCCGGUUUUCCCCCAAUCUUUUCAAGUUAUCAAACAAUAAAAGUAAAUAAAAAUGUUUUCAAAACAAAAAACAUUCUCUUCUU----------UUGGACCAAACAAA ............((((((.......))........................(((((........((((((((........)))))))).....)))----------))))))........ ( -10.92) >DroSec_CAF1 3502 108 - 1 AACACUUUCUUGGUCCGGUUU--CCCCAACCUUUUCAAGUUAUCAAACAAUAAAAGUAAAUAAAAAUGUUUUCAAAACAAAAAACAUUCUCUUCUU----------UUGGACCAAACUAA .........(((((((((((.--....))))....................(((((........((((((((........)))))))).....)))----------)))))))))..... ( -18.62) >DroSim_CAF1 3815 108 - 1 AACACUUUCUUGGUCCGGUUU--CCCCAACCUUUUCAAGUUAUCAAACAAUAAAAGUAAAUAAAAAUGUUUUCAAAACAAAAAACAUUCUCUUCUU----------UUGGACGAAACUAA .........((.((((((((.--....))))....................(((((........((((((((........)))))))).....)))----------)))))).))..... ( -14.22) >DroEre_CAF1 3462 117 - 1 AAUACUUUCUUGGUCCGGUUU--CCCCAACCUUUUCAAGUUAUCACACAAUAAAAGUAACUAA-AGUGUUUUCAAAACAAAACGCAUUCUUUUCUUUGUCACCGGUUUGGACCAAACUAA .........(((((((((...--..))((((......((((((............))))))..-.(((((((......)))))))..................)))).)))))))..... ( -22.90) >DroYak_CAF1 3648 118 - 1 AACAAUUUCUUGGUCCGGUUU--CACCAACCUUUUAAAGUUGUCAUACAAUAAAAGUAACUAAAAAUGUUUCCAAAACAAAACGCAUUCUUCUCUUUGUCACCGUUUUGGACCAAACUAA .........(((((((((...--..))...((((((..((......))..))))))......................((((((....(........)....)))))))))))))..... ( -18.10) >consensus AACACUUUCUUGGUCCGGUUU__CCCCAACCUUUUCAAGUUAUCAAACAAUAAAAGUAAAUAAAAAUGUUUUCAAAACAAAAAACAUUCUCUUCUU__________UUGGACCAAACUAA .........(((((((((((.......)))).................................((((((((........))))))))....................)))))))..... (-13.30 = -13.74 + 0.44)


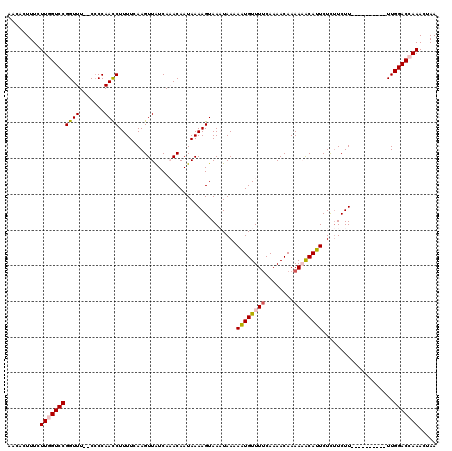
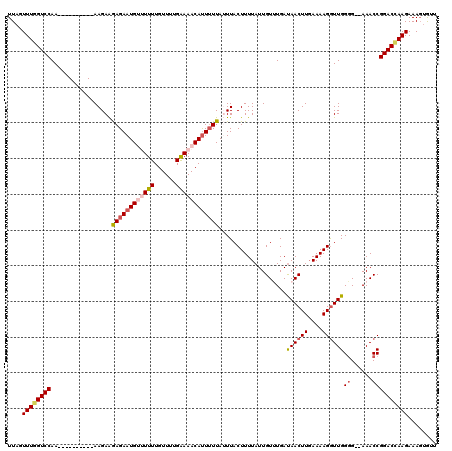
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:30 2006