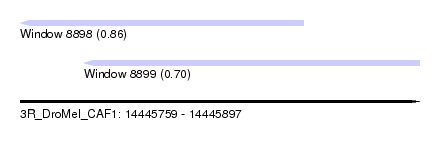
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,445,759 – 14,445,897 |
| Length | 138 |
| Max. P | 0.858498 |

| Location | 14,445,759 – 14,445,857 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 89.51 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.35 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
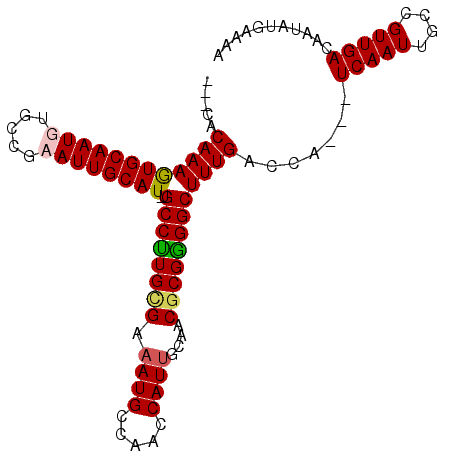
>3R_DroMel_CAF1 14445759 98 - 27905053 ---CACAAAGUGCAAUGUGCCGCAUUGCAU-UGCCUUGCGAAAUGCCAACCAUUUCAACGCGGGGCUUUGACCAUCAUCAAUUGCCGUUGACAAUAUGAAAA ---..((((((((((((.....))))))))-)(((((((((((((.....))))))...))))))).)))....((((..((((.......))))))))... ( -27.60) >DroSec_CAF1 2897 99 - 1 CUCCACAAAGUGCAAUGUGCCGCAUUGCAUUUGCCUUGCGAAAUGCCAAGCAUUUCAACGCGAGGCUUUGACCA---UCAAUUGCCGUUGACAAUAUGAAAA .......((((((((((.....))))))))))((((((((((((((...)))))))...)))))))........---(((((....)))))........... ( -31.30) >DroEre_CAF1 2882 95 - 1 ---CUCAAAAUGCAAUGUGCCGAAUUGCAU-UGCCUUGUGAAAUGCCAACCAUGGCGACGCGGGGCUUUGACCA---UCAAUUGCCGUUGACAAUAUGAAAA ---.((((((((((((.......)))))))-)((((((((...(((((....))))).)))))))).))))...---(((((....)))))........... ( -27.90) >DroYak_CAF1 3042 95 - 1 ---CUCAAAGUGCAAUGUGCCGAAUUGCAU-GGCCCUGUGAAAUGCCAACCAUUGCGACCCGGGGCUUUGACCA---UCAAUUGCCGUUGACAAUAUGAAAA ---.(((...(((((((.((..(((((((.-(((((((.(...(((........)))..)))))))).))....---.)))))))))))).))...)))... ( -23.20) >consensus ___CACAAAGUGCAAUGUGCCGAAUUGCAU_UGCCUUGCGAAAUGCCAACCAUUGCAACGCGGGGCUUUGACCA___UCAAUUGCCGUUGACAAUAUGAAAA .....((((((((((((.....))))))))..((((((((.((((.....))))....)))))))))))).......(((((....)))))........... (-22.04 = -22.35 + 0.31)


| Location | 14,445,781 – 14,445,897 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.37 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -20.66 |
| Energy contribution | -20.97 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14445781 116 - 27905053 CUUAUGCAAUCCCUGAAUCCACCAACAACAGCUCCCCCAC---CACAAAGUGCAAUGUGCCGCAUUGCAU-UGCCUUGCGAAAUGCCAACCAUUUCAACGCGGGGCUUUGACCAUCAUCA .............(((........................---.....(((((((((.....))))))))-)(((((((((((((.....))))))...)))))))........)))... ( -26.10) >DroSec_CAF1 2919 115 - 1 CUUAUGCAAUCCCUGAAUCCACCAA--ACAGCUCUCCCACCUCCACAAAGUGCAAUGUGCCGCAUUGCAUUUGCCUUGCGAAAUGCCAAGCAUUUCAACGCGAGGCUUUGACCA---UCA ......................(((--(...................((((((((((.....))))))))))((((((((((((((...)))))))...)))))))))))....---... ( -30.50) >DroEre_CAF1 2904 107 - 1 CUUAUGCAAUCCCUGAAUCCACCA---ACAGCUCCCC------CUCAAAAUGCAAUGUGCCGAAUUGCAU-UGCCUUGUGAAAUGCCAACCAUGGCGACGCGGGGCUUUGACCA---UCA ........................---..........------.((((((((((((.......)))))))-)((((((((...(((((....))))).)))))))).))))...---... ( -26.30) >DroYak_CAF1 3064 107 - 1 CUUAUGCAAUCCCUGAAUCCACCA---ACAGCUCCCC------CUCAAAGUGCAAUGUGCCGAAUUGCAU-GGCCCUGUGAAAUGCCAACCAUUGCGACCCGGGGCUUUGACCA---UCA ......................((---(.((((((..------.......((((((((((......)))(-(((..........))))..)))))))....)))))))))....---... ( -21.22) >consensus CUUAUGCAAUCCCUGAAUCCACCA___ACAGCUCCCC______CACAAAGUGCAAUGUGCCGAAUUGCAU_UGCCUUGCGAAAUGCCAACCAUUGCAACGCGGGGCUUUGACCA___UCA .............................................((((((((((((.....))))))))..((((((((.((((.....))))....)))))))))))).......... (-20.66 = -20.97 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:28 2006