
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,444,851 – 14,445,147 |
| Length | 296 |
| Max. P | 0.982965 |

| Location | 14,444,851 – 14,444,971 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.22 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -23.36 |
| Energy contribution | -22.40 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14444851 120 - 27905053 GAAUUCCAAGCGCUGCAUCCAUGUAAAAAAUACCUGGAGCGGGCCGAGUUCGUCCGAAAUAUUCCAAAAUGCAUAGCGCACGGCAGAGGACGGGAAAGAAAUGAACCCCAUUCCUGAAAA .....((..((((((((((((.(((.....))).)))).(((((.(....)))))).............)))..)))))..))(((.(((.(((..(....)....))).)))))).... ( -31.30) >DroSec_CAF1 2003 118 - 1 GAAUUCCAAGCGCUGCAUCCGUGUAAAAAAUACCUGGAGCGGGUCGAGUUCGUCCGAAAUAUUCCAAAAUGCAAAGCGCACGGCAGAGGAUGAGAAAGAAAUGAAU--CAUUCCUGCAAA .........((((((((((((.(((.....))).)))).((((.((....)))))).............)))..)))))...((((.((((((...(....)...)--)))))))))... ( -35.60) >DroSim_CAF1 2072 118 - 1 GAAUUCCAAGCGCUGCAUCCAUGUAAAAAAUACCUGGAGCGGGUCGCGUUCGUCCGAAAUAUUCCAAAAUGCAUAGCGCACGGCAGAGGAUGAGAAAGAAAUGAAU--CAUUCCUGCAAA .........(((((((.((((.(((.....))).))))))(((.....(((....)))....))).........)))))...((((.((((((...(....)...)--)))))))))... ( -34.80) >DroEre_CAF1 1973 116 - 1 GAAUUCCAAGUGCUGCAUCCAUGUAAAAAAUACCUGGAGCGGGCAGAGUUCGUCCGAAAUAUUCCGAACUGCAUAA--CACGGCAGUGGAUGAGAAAGAAAUGAAC--CAUCCCUGAAAA .........(((((((.((((.(((.....))).))))))))(((..(((((............))))))))....--)))..(((.(((((....(....)....--)))))))).... ( -29.70) >DroYak_CAF1 2105 116 - 1 GAAUUCCAAGUGCUGCAUCUAUGUAAAAAAUACCUGGAGCGGACAGAGUUCGUCCGAAAUAUUCCAAACUGCAAGA--CACGGCGGAGGAUGGGAAAGAAAUGAAC--CAUCCCUGAAAA .........((((((((((((.(((.....))).)))).(((((.......))))).............))).)).--)))..(((.((((((...(....)...)--)))))))).... ( -31.20) >consensus GAAUUCCAAGCGCUGCAUCCAUGUAAAAAAUACCUGGAGCGGGCCGAGUUCGUCCGAAAUAUUCCAAAAUGCAUAGCGCACGGCAGAGGAUGAGAAAGAAAUGAAC__CAUUCCUGAAAA ((((((...((.((((.((((.(((.....))).)))))))))).))))))................................(((.(((((....(....)......)))))))).... (-23.36 = -22.40 + -0.96)

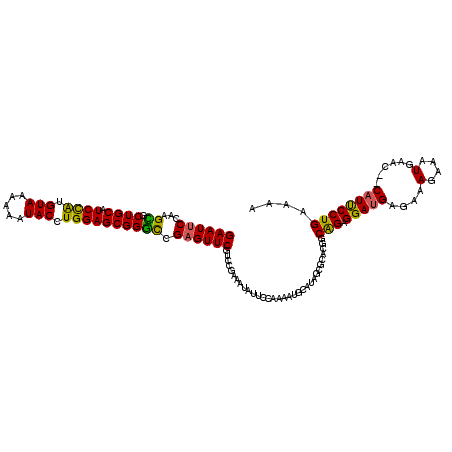

| Location | 14,444,931 – 14,445,051 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -31.60 |
| Energy contribution | -31.68 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14444931 120 - 27905053 CAUCGGGGGAGCAGCACAGCAGAAAGGAAUGUGUGCAUUUGUCUGGCACUGACUUCGUGGGAGAUUGGGCAGAUGUUUUGGAAUUCCAAGCGCUGCAUCCAUGUAAAAAAUACCUGGAGC ..(((((((((((((.(....)...(((((....(((((((((..((.((.((...)).)).).)..)))))))))......)))))....))))).))).(((.....))))))))... ( -36.30) >DroSec_CAF1 2081 117 - 1 CAACGGUGGAG---CGCAGCAGCAAGGAAUGUGUGCAUUUGUCUGGCACUGACUUCGUGGGAGAUUGGGCAGAUGUUUUGGAAUUCCAAGCGCUGCAUCCGUGUAAAAAAUACCUGGAGC ...(((.(((.---.(((((.((..(((((....(((((((((..((.((.((...)).)).).)..)))))))))......)))))..))))))).)))((((.....))))))).... ( -39.00) >DroSim_CAF1 2150 117 - 1 CAACGGGGGAG---CGCAGCAGCAAGGAAUGUGUGCAUUUGUCUGGCACUGACUUCGUGGGAGAUUGGGCAGAUGUUUUGGAAUUCCAAGCGCUGCAUCCAUGUAAAAAAUACCUGGAGC ...(((((((.---.(((((.((..(((((....(((((((((..((.((.((...)).)).).)..)))))))))......)))))..))))))).))).(((.....))))))).... ( -40.20) >DroEre_CAF1 2049 112 - 1 C---GGGGGA-----GCAGCAGAAAGGAAUGUGUGCAUUUGUCUGGCACUGACUUCGUGGGAGAUUGGGCAGAUGUUUCGGAAUUCCAAGUGCUGCAUCCAUGUAAAAAAUACCUGGAGC (---((((((-----((((((....(((((.((.(((((((((..((.((.((...)).)).).)..)))))))))..))..)))))...)))))).))).(((.....))))))).... ( -37.60) >DroYak_CAF1 2181 112 - 1 C---GGGGGA-----ACAGCAGAAAGGAAUGUGUGCAUUUGUCUGGCACUGACUUCGUGGGAGAUUGGGCAGAUGUUUCGGAAUUCCAAGUGCUGCAUCUAUGUAAAAAAUACCUGGAGC (---((((((-----.(((((....(((((.((.(((((((((..((.((.((...)).)).).)..)))))))))..))..)))))...)))))..))).(((.....))))))).... ( -30.70) >consensus CA_CGGGGGAG___CGCAGCAGAAAGGAAUGUGUGCAUUUGUCUGGCACUGACUUCGUGGGAGAUUGGGCAGAUGUUUUGGAAUUCCAAGCGCUGCAUCCAUGUAAAAAAUACCUGGAGC ...............(((((.....(((((....(((((((((..((.((.((...)).)).).)..)))))))))......)))))....))))).((((.(((.....))).)))).. (-31.60 = -31.68 + 0.08)



| Location | 14,445,011 – 14,445,131 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.71 |
| Mean single sequence MFE | -40.94 |
| Consensus MFE | -34.32 |
| Energy contribution | -34.36 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14445011 120 - 27905053 UUGGAGCCGUGGGGCAAAGUUGAGGUAAACGUGCUCCACAUCGAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAUCAUCGGGGGAGCAGCACAGCAGAAAGGAAUGUGUGCAUUU ........((((((((..(((......))).))))))))...(((((((((.(((((((.(((....(((..(..(((......)))..)))).))).)))......))))))))))))) ( -41.90) >DroSec_CAF1 2161 117 - 1 UUGGAGCCGUGGGGCAAAGUUGAGGUAAACGUGCUCCACAUCGAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAUCAACGGUGGAG---CGCAGCAGCAAGGAAUGUGUGCAUUU ........((((((((..(((......))).))))))))...(((((((((((((((.......)))))).(((((((((.....)))).(---(......))..))))).))))))))) ( -38.80) >DroSim_CAF1 2230 117 - 1 UUGGAGCCGUGGGGCAAAGUUGAGGUAAACGUGCUCCACAUCGAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAUCAACGGGGGAG---CGCAGCAGCAAGGAAUGUGUGCAUUU ........((((((((..(((......))).))))))))...(((((((((.(((((....((....(((..(..(((......)))..).---.)))....))..)))))))))))))) ( -41.90) >DroEre_CAF1 2129 112 - 1 UUGGAGCCGUGGGCCAAAGUUGAGGUAAACGUGCUCCACAUCGAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAUC---GGGGGA-----GCAGCAGAAAGGAAUGUGUGCAUUU .(((((((((..(((........)))..))).))))))....(((((((((.((((((((.......)))..((((((...---.)))))-----)..........)))))))))))))) ( -41.10) >DroYak_CAF1 2261 112 - 1 UUGGAGCCGUGGGCCACAGUUGAGGUAAACGUGCUCCACAUCAAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAUC---GGGGGA-----ACAGCAGAAAGGAAUGUGUGCAUUU .(((((((((..(((........)))..))).))))))....(((((((((.((((((((.......)))..((((((...---.)))))-----)..........)))))))))))))) ( -41.00) >consensus UUGGAGCCGUGGGGCAAAGUUGAGGUAAACGUGCUCCACAUCGAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAUCA_CGGGGGAG___CGCAGCAGAAAGGAAUGUGUGCAUUU .(((((((((..(.(........).)..))).))))))....(((((((((.((((((((.......)))..((((((......))))))................)))))))))))))) (-34.32 = -34.36 + 0.04)

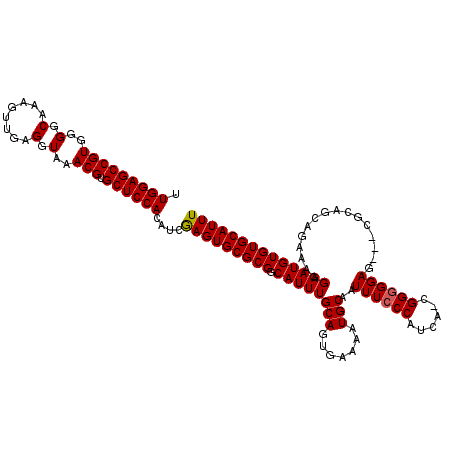

| Location | 14,445,051 – 14,445,147 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.61 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -28.14 |
| Energy contribution | -30.02 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14445051 96 - 27905053 GAGUAGG------------------------GAUUCAUAGUUGGAGCCGUGGGGCAAAGUUGAGGUAAACGUGCUCCACAUCGAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAU .....((------------------------((((((....))))(((((((((((..(((......))).))))))))..((....)).)))..(((((.......)))))..)))).. ( -31.10) >DroSec_CAF1 2198 120 - 1 GAGUAGGGAGUGCGGACGGAGGGAAUACGGGGAUUCAUAGUUGGAGCCGUGGGGCAAAGUUGAGGUAAACGUGCUCCACAUCGAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAU .....(((((((((..(((...........((.((((....)))).))((((((((..(((......))).)))))))).)))..))))).((((((.......))))))....)))).. ( -41.10) >DroSim_CAF1 2267 120 - 1 GAGUAGGGAGUGCGGACGGAGGGAAUACGGGGAUUCAUAGUUGGAGCCGUGGGGCAAAGUUGAGGUAAACGUGCUCCACAUCGAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAU .....(((((((((..(((...........((.((((....)))).))((((((((..(((......))).)))))))).)))..))))).((((((.......))))))....)))).. ( -41.10) >DroEre_CAF1 2161 118 - 1 GAGUAGGGAGUGCGGACGGAGGGAAUACG--GAUUCAUAGUUGGAGCCGUGGGCCAAAGUUGAGGUAAACGUGCUCCACAUCGAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAU .....(((((((((..((..(.((((...--.)))).....(((((((((..(((........)))..))).)))))))..))..))))).((((((.......))))))....)))).. ( -41.40) >DroYak_CAF1 2293 118 - 1 GAGAAGGGAGUGCGGGCGGAGGGAAAACG--GAUUCAUAGUUGGAGCCGUGGGCCACAGUUGAGGUAAACGUGCUCCACAUCAAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAU .....(((((((((.((.....(((....--..)))...((.((((((((..(((........)))..))).))))))).....)).))))((((((.......))))))...))))).. ( -39.90) >consensus GAGUAGGGAGUGCGGACGGAGGGAAUACG__GAUUCAUAGUUGGAGCCGUGGGGCAAAGUUGAGGUAAACGUGCUCCACAUCGAGUGCGCGGCAUUUGCAGUGAAAAUGCAAUUUCCCAU .....(((((.(((.((................((((....))))...((((((((..(((......))).)))))))).....)).))).((((((.......))))))...))))).. (-28.14 = -30.02 + 1.88)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:22 2006