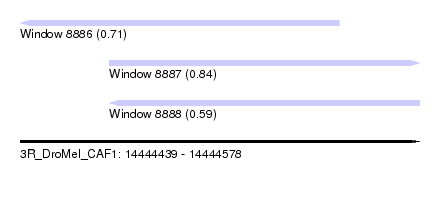
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,444,439 – 14,444,578 |
| Length | 139 |
| Max. P | 0.835238 |

| Location | 14,444,439 – 14,444,550 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -20.96 |
| Consensus MFE | -14.81 |
| Energy contribution | -15.09 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14444439 111 - 27905053 GAUUUUUCACUUCCCAUACGGAAUGUAAGUAGAAAAAAAGCAGCAAAAGAGCGGGAGUUUUAAGUUUGGGGCGUGAAAGGAAAAAGUAAAAUGUUUGCCAAAA---------AAAAAAGU ..((((((((.(((((.((.........((.(........).))...(((((....)))))..)).))))).)))))))).....((((.....)))).....---------........ ( -20.00) >DroSec_CAF1 1607 113 - 1 GAUUUUUCACUUCCCAUGUGCAGCGUAAG----AAAAAAGCAGCAAAAGAGCGGGAGUUUUGGGUUUGGGGCGUGAAAGGAAAAAGUAAAAUGUUUGCCAAAAUAAAA---AACAAAAGU ..((((((((.(((((..(((.((.....----......)).)))..(((((....))))).....))))).))))))))...........(((((...........)---))))..... ( -24.00) >DroSim_CAF1 1678 116 - 1 GAUUUUUCACUUCCCAUAUGCAGCGUAAG----AAAAAAGCAGCAAAAGAGCGGGAGUUUUGGGUUUGGGGCGUGAAAGGAAAAAGUAAAAUGUUUGCCAAAAUAAAAAGAAAAAAAAGU ..((((((((.(((((..(((.((.....----......)).)))..(((((....))))).....))))).))))))))...........(((((....)))))............... ( -23.20) >DroEre_CAF1 1593 97 - 1 GAUUUUUCACUUCCCGUGUGGAG---AGG----AAAAAAGCAGCAAAGGAGCGGGAGUU-UGAGUUUGGGGCGUGAAAGGAAAAAGUAAAAUGCUUGCCAAAAAA--------------- ....(((((((((((.......)---.))----))....((..((((.((((....)))-)...))))..))))))))((...((((.....)))).))......--------------- ( -20.70) >DroYak_CAF1 1705 105 - 1 GAUUUUUCACUUCCAAU--AAAG---AGA----AAAAAAUCAGCCAAAGAGCGGGAGUU-UGAGUUUGGGGCGUGAAAGGAA-AAGUAAAAUGUUUGCCAAAAAA-AA---AACAAAAAU ..((((((((((((((.--...(---(..----......)).......((((....)))-)....)))))).))))))))..-........(((((.........-.)---))))..... ( -16.90) >consensus GAUUUUUCACUUCCCAUAUGCAG_GUAAG____AAAAAAGCAGCAAAAGAGCGGGAGUUUUGAGUUUGGGGCGUGAAAGGAAAAAGUAAAAUGUUUGCCAAAAAA_AA___AAAAAAAGU ..((((((((.(((((.........................(((...(((((....)))))..)))))))).)))))))).....((((.....))))...................... (-14.81 = -15.09 + 0.28)

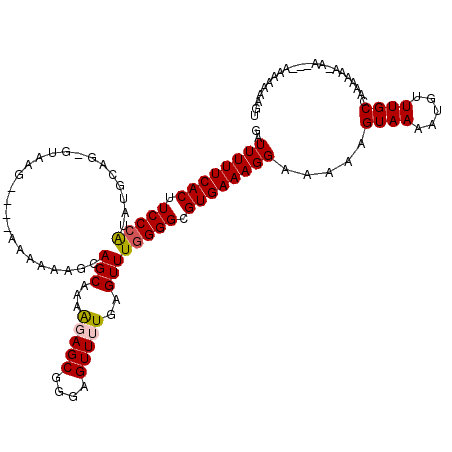
| Location | 14,444,470 – 14,444,578 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.52 |
| Mean single sequence MFE | -16.50 |
| Consensus MFE | -10.38 |
| Energy contribution | -10.57 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14444470 108 + 27905053 CCUUUCACGCCCCAAACUUAAAACUCCCGCUCUUUUGCUGCUUUUUUUCUACUUACAUUCCGUAUGGGAAGUGAAAAAUCUUGUUUUCUUG------------UAUAUAUGUAUGUAUGC ..((((((..((((..............((......))...............(((.....)))))))..))))))..............(------------((((((....))))))) ( -17.10) >DroSec_CAF1 1644 85 + 1 CCUUUCACGCCCCAAACCCAAAACUCCCGCUCUUUUGCUGCUUUUUU----CUUACGCUGCACAUGGGAAGUGAAAAAUCUUGUUUUC-------------------------------C ..((((((..((((..............((......))(((......----........)))..))))..))))))............-------------------------------. ( -15.14) >DroSim_CAF1 1718 85 + 1 CCUUUCACGCCCCAAACCCAAAACUCCCGCUCUUUUGCUGCUUUUUU----CUUACGCUGCAUAUGGGAAGUGAAAAAUCUUGUUUUC-------------------------------C ..((((((..((((..............((......))(((......----........)))..))))..))))))............-------------------------------. ( -15.24) >DroYak_CAF1 1740 110 + 1 CCUUUCACGCCCCAAACUCA-AACUCCCGCUCUUUGGCUGAUUUUUU----UCU---CUUU--AUUGGAAGUGAAAAAUCUUGUUUUCCCGAGCAUAUAUAUGUAUAUAUGUAUAUCCGC ........((..........-.......((((...((..(((.((((----((.---((((--....)))).))))))....)))..)).))))(((((((((....)))))))))..)) ( -18.50) >consensus CCUUUCACGCCCCAAACCCAAAACUCCCGCUCUUUUGCUGCUUUUUU____CUUACGCUGCAUAUGGGAAGUGAAAAAUCUUGUUUUC_______________________________C ..((((((..((((..............((......))(((..................)))..))))..))))))............................................ (-10.38 = -10.57 + 0.19)

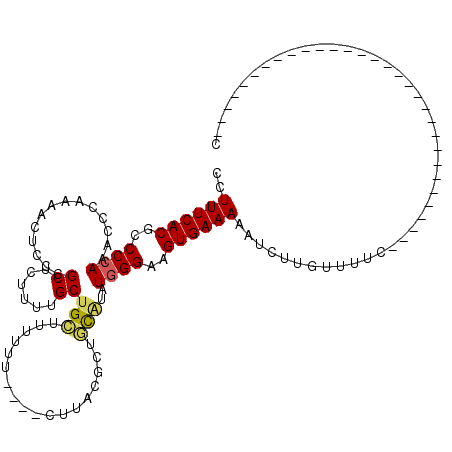
| Location | 14,444,470 – 14,444,578 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.52 |
| Mean single sequence MFE | -20.65 |
| Consensus MFE | -14.06 |
| Energy contribution | -14.56 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
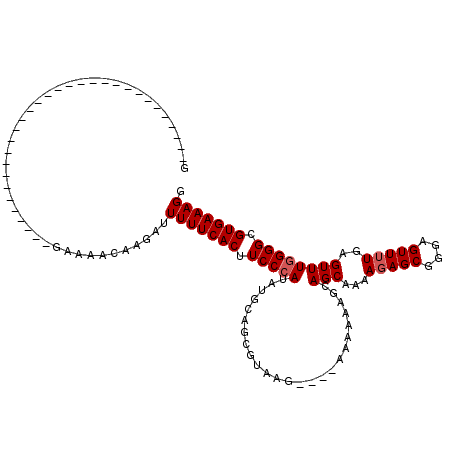
>3R_DroMel_CAF1 14444470 108 - 27905053 GCAUACAUACAUAUAUA------------CAAGAAAACAAGAUUUUUCACUUCCCAUACGGAAUGUAAGUAGAAAAAAAGCAGCAAAAGAGCGGGAGUUUUAAGUUUGGGGCGUGAAAGG .................------------..............(((((((.(((((.((.........((.(........).))...(((((....)))))..)).))))).))))))). ( -19.30) >DroSec_CAF1 1644 85 - 1 G-------------------------------GAAAACAAGAUUUUUCACUUCCCAUGUGCAGCGUAAG----AAAAAAGCAGCAAAAGAGCGGGAGUUUUGGGUUUGGGGCGUGAAAGG .-------------------------------...........(((((((.(((((..(((.((.....----......)).)))..(((((....))))).....))))).))))))). ( -21.90) >DroSim_CAF1 1718 85 - 1 G-------------------------------GAAAACAAGAUUUUUCACUUCCCAUAUGCAGCGUAAG----AAAAAAGCAGCAAAAGAGCGGGAGUUUUGGGUUUGGGGCGUGAAAGG .-------------------------------...........(((((((.(((((..(((.((.....----......)).)))..(((((....))))).....))))).))))))). ( -22.00) >DroYak_CAF1 1740 110 - 1 GCGGAUAUACAUAUAUACAUAUAUAUGCUCGGGAAAACAAGAUUUUUCACUUCCAAU--AAAG---AGA----AAAAAAUCAGCCAAAGAGCGGGAGUU-UGAGUUUGGGGCGUGAAAGG .......................(((((((.(((...(((((....))((((((...--...(---(..----......)).((......)))))))))-))..))).)))))))..... ( -19.40) >consensus G_______________________________GAAAACAAGAUUUUUCACUUCCCAUAUGCAGCGUAAG____AAAAAAGCAGCAAAAGAGCGGGAGUUUUGAGUUUGGGGCGUGAAAGG ...........................................(((((((.(((((.........................(((...(((((....)))))..)))))))).))))))). (-14.06 = -14.56 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:17 2006