
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
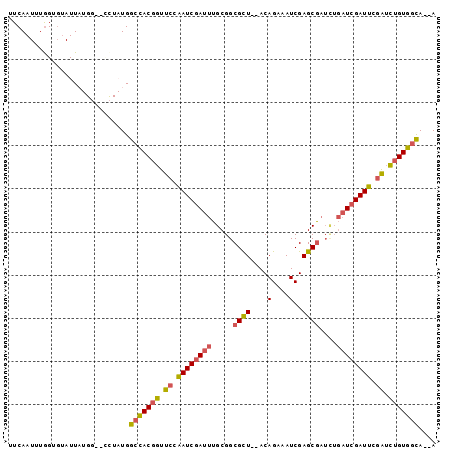
| Location | 14,432,013 – 14,432,106 |
| Length | 93 |
| Max. P | 0.832521 |

| Location | 14,432,013 – 14,432,106 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.03 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -22.50 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14432013 93 + 27905053 UUCAAUUUGGCGUAUUAUGG--CCUAUGGCCACGGUUCCAAUCGAUUUGCGGCGCU--ACAGAAAUCGAGCGAUCUGAUCGAUUCGAUCUGUGGCA--A ........(.((((...(((--((...)))))((((....))))...)))).)(((--(((((..(((((((((...)))).))))))))))))).--. ( -33.40) >DroSec_CAF1 210463 95 + 1 UUCAAUUUGGUGUAUUUUGG--CCUAUGGCCACGGUUCCAAUCGAUUUGCGGCGCUAUACAGAAAUCGAGCGAUCUGAUCGAUUCGAUCUGUGGCA--A ........(((........)--))....(((((((.((.((((((((.....((((....(....)..))))....)))))))).)).))))))).--. ( -30.10) >DroSim_CAF1 202660 95 + 1 UUCAAUUUGGUGUAUUAUGG--CCUAUGGCCACGGUUCCAAUCGAUUUGCGGCGCUAUACAGAAAUCGAGCGAUCUGAUCGAUUCGAUCUGUGGCA--A ........(((........)--))....(((((((.((.((((((((.....((((....(....)..))))....)))))))).)).))))))).--. ( -30.10) >DroEre_CAF1 218826 86 + 1 UUCAAUUUGGUGUAUUA---------UGGCCACGGUUCGAAUCCAUUUGCGGCGUU--ACAGAAAUCGAGCGAUCUGAUCGAUUCGAUCUGUGGCA--A .................---------..(((((((.(((((((.(((.....((((--...(....).))))....))).))))))).))))))).--. ( -27.10) >DroYak_CAF1 221603 86 + 1 UUCAAUUUGGUGUAUUA---------UGGCCACGGUUCCAAUCGAUUUGCGGCGCU--ACAGAAAUCGAGCGCUUUGAUCGAUUCGAUCUGUGGCA--A .................---------..(((((((.((.((((((((...((((((--...(....).))))))..)))))))).)).))))))).--. ( -33.80) >DroAna_CAF1 195363 89 + 1 UUCAAUUUAG---AUUUUGCUUUUUAUGGCUACGGUUCCGAUCGAUUUGCGGCGCU--ACAGAAAUCGAGCAA-----UCGAUUUAGAUAGUAAUAGUA ..........---....((((.......((..(((.((.....)).)))..))(((--(...(((((((....-----)))))))...))))...)))) ( -16.30) >consensus UUCAAUUUGGUGUAUUAUGG__CCUAUGGCCACGGUUCCAAUCGAUUUGCGGCGCU__ACAGAAAUCGAGCGAUCUGAUCGAUUCGAUCUGUGGCA__A ............................(((((((.((.((((((((.....((((.....(....).))))....)))))))).)).))))))).... (-22.50 = -22.70 + 0.20)


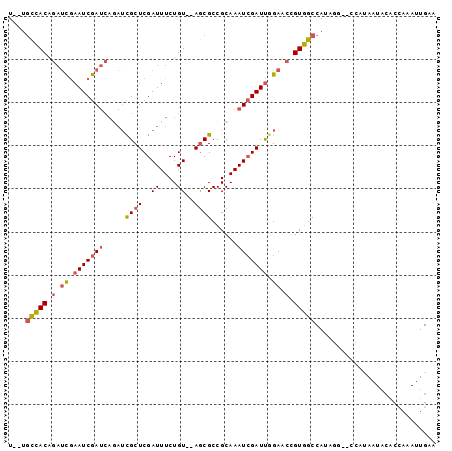
| Location | 14,432,013 – 14,432,106 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.03 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -16.01 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14432013 93 - 27905053 U--UGCCACAGAUCGAAUCGAUCAGAUCGCUCGAUUUCUGU--AGCGCCGCAAAUCGAUUGGAACCGUGGCCAUAGG--CCAUAAUACGCCAAAUUGAA .--.(((((.(.((.(((((((..(..((((((.....)).--))))...)..))))))).)).).)))))....((--(........)))........ ( -27.90) >DroSec_CAF1 210463 95 - 1 U--UGCCACAGAUCGAAUCGAUCAGAUCGCUCGAUUUCUGUAUAGCGCCGCAAAUCGAUUGGAACCGUGGCCAUAGG--CCAAAAUACACCAAAUUGAA .--.(((((.(.((.(((((((..(..((((..((....))..))))...)..))))))).)).).)))))......--.................... ( -23.30) >DroSim_CAF1 202660 95 - 1 U--UGCCACAGAUCGAAUCGAUCAGAUCGCUCGAUUUCUGUAUAGCGCCGCAAAUCGAUUGGAACCGUGGCCAUAGG--CCAUAAUACACCAAAUUGAA .--.((.(((((((((..((((...)))).))))..)))))...))........((((((((....((((((...))--))))......)).)))))). ( -23.50) >DroEre_CAF1 218826 86 - 1 U--UGCCACAGAUCGAAUCGAUCAGAUCGCUCGAUUUCUGU--AACGCCGCAAAUGGAUUCGAACCGUGGCCA---------UAAUACACCAAAUUGAA .--.(((((.(.(((((((.((.(((((....))))).(((--......))).)).))))))).).)))))..---------................. ( -20.90) >DroYak_CAF1 221603 86 - 1 U--UGCCACAGAUCGAAUCGAUCAAAGCGCUCGAUUUCUGU--AGCGCCGCAAAUCGAUUGGAACCGUGGCCA---------UAAUACACCAAAUUGAA .--.(((((.(.((.(((((((....(((((((.....)).--))))).....))))))).)).).)))))..---------................. ( -26.60) >DroAna_CAF1 195363 89 - 1 UACUAUUACUAUCUAAAUCGA-----UUGCUCGAUUUCUGU--AGCGCCGCAAAUCGAUCGGAACCGUAGCCAUAAAAAGCAAAAU---CUAAAUUGAA ........((((..(((((((-----....)))))))..))--)).........(((((((....))..((........)).....---....))))). ( -11.50) >consensus U__UGCCACAGAUCGAAUCGAUCAGAUCGCUCGAUUUCUGU__AGCGCCGCAAAUCGAUUGGAACCGUGGCCAUAGG__CCAUAAUACACCAAAUUGAA ....(((((.(.((.(((((((.....((((..((....))..))))......))))))).)).).)))))............................ (-16.01 = -16.48 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:14 2006