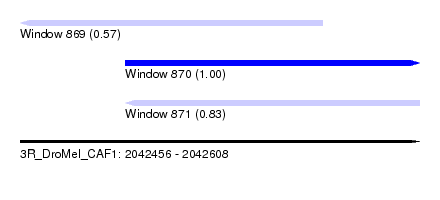
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,042,456 – 2,042,608 |
| Length | 152 |
| Max. P | 0.996324 |

| Location | 2,042,456 – 2,042,571 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.89 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -16.05 |
| Energy contribution | -16.86 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
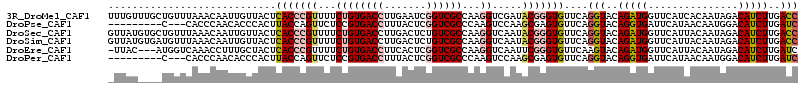
>3R_DroMel_CAF1 2042456 115 - 27905053 UUUGUUUGCUGUUUAAACAAUUGUUACUCACCCGUUUUCUGUGACCUUGAAUCGGUCGCCAAGGUCGAUACGGGUGUUCAGGUACAGAUGGUUCAUCACAAUAGACAUCUUGACC ..((((((.(((...(((..((((.((((((((((......((((((((..........))))))))..)))))))....)))))))...)))....))).))))))........ ( -30.80) >DroPse_CAF1 163287 103 - 1 ---------C---CACCCAACACCCACUUACCAGUUCUCCGUGACCUUUACUCGGUCGCCCAAGUCCAAGCGAGUGUUCAGGUACAGGUGAUUCAUAACAAUGGACAUCUUGAUC ---------.---.(((.(((((.(.(((....(..((..((((((.......))))))...))..)))).).)))))..)))..(((((.(((((....))))))))))..... ( -22.80) >DroSec_CAF1 185642 115 - 1 GUUAUGUGCUGUUUAAACAAUUGUUACUCACCCGUUUUCUGUGACCUUGACUCUGUCGCCAAGGUCAAUACGGGUGUUCAGGUACAGAUGGUUCAUUACAAUAGACAUCUUGACC ((((((((((.....(((....)))...(((((((......((((((((.(......).))))))))..)))))))....))))))((((...............)))).)))). ( -32.66) >DroSim_CAF1 190546 115 - 1 GUUAUGUGAUGUUUAAACAAUUGUUACUCACCCGUUUUCUGUGACCUUGACUCUGUCGCCAAGGUCAAUACGGGUGUUCAGGUACAGAUGGUUCAUUACAAUAGACAUCUUGACC ((((...((((((((..((.((((.((((((((((......((((((((.(......).))))))))..)))))))....))))))).))((.....))..)))))))).)))). ( -36.50) >DroEre_CAF1 185573 111 - 1 -UUAC---AUGGUCAAACCUUUGCUACUCACCCGUUUUCUGUGACCUUCACUCGGUCGCCAAGGUCAAUUCGGGUGUUCAAGUACAGAUGGUUCAUUACAAUAGACAUCUUGAUC -....---..(((((((((((((.((((((((((...(((((((((.......))))))..)))......))))))....)))))))).)))).........((....))))))) ( -27.90) >DroPer_CAF1 164898 103 - 1 ---------C---CACCCAACACCCACUUACCAGUUCUCCGUGACCUUUACUCGGUCGCCCAAGUCCAAGCGAGUGUUCAGGUACAGGUGAUUCAUAACAAUGGACAUCUUGAUC ---------.---.(((.(((((.(.(((....(..((..((((((.......))))))...))..)))).).)))))..)))..(((((.(((((....))))))))))..... ( -22.80) >consensus _UUAU____UGUUCAAACAAUUGCUACUCACCCGUUUUCUGUGACCUUGACUCGGUCGCCAAGGUCAAUACGGGUGUUCAGGUACAGAUGGUUCAUUACAAUAGACAUCUUGACC ............................(((((((...((((((((.......))))))...)).....)))))))....(((..(((((...............)))))..))) (-16.05 = -16.86 + 0.81)
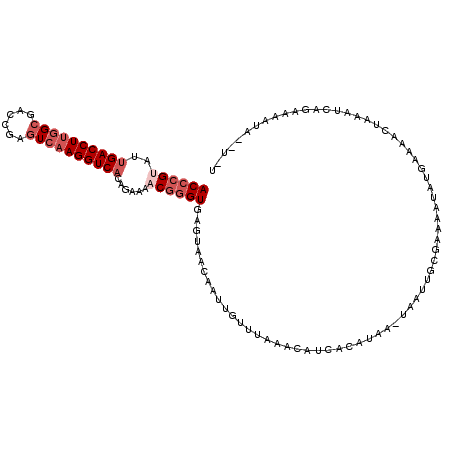
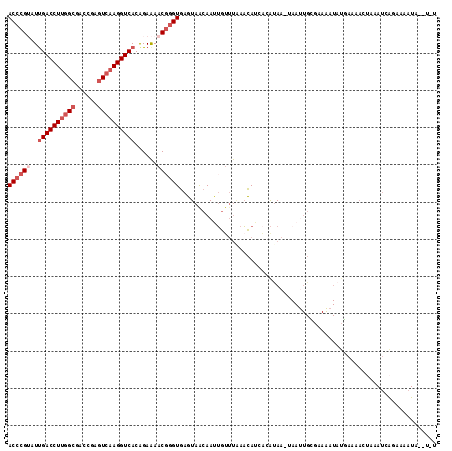
| Location | 2,042,496 – 2,042,608 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.16 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -16.50 |
| Energy contribution | -18.10 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
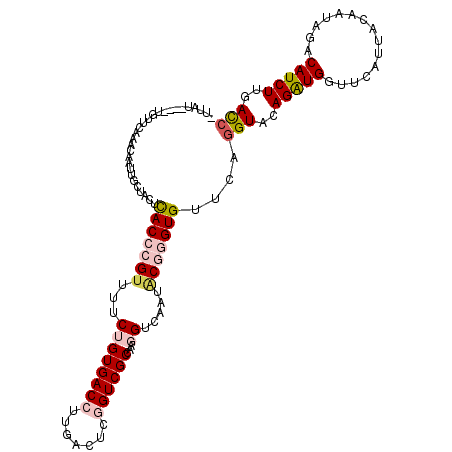
>3R_DroMel_CAF1 2042496 112 + 27905053 ACCCGUAUCGACCUUGGCGACCGAUUCAAGGUCACAGAAAACGGGUGAGUAACAAUUGUUUAAACAGCAAACAAAUAAUUGAGAAAAUAUUAAAACAAAAUCGGAAAAUA--U-U ((((((.((((((((((........))))))))...))..))))))......(((((((((...........))))))))).............................--.-. ( -22.50) >DroSec_CAF1 185682 112 + 1 ACCCGUAUUGACCUUGGCGACAGAGUCAAGGUCACAGAAAACGGGUGAGUAACAAUUGUUUAAACAGCACAUAACUAAUUGCGAAAAUAUGAAAACUAAAUCAGAAAAUA--U-U ((((((..((((((((((......))))))))))......))))))......((((((((............))).))))).............................--.-. ( -25.20) >DroSim_CAF1 190586 112 + 1 ACCCGUAUUGACCUUGGCGACAGAGUCAAGGUCACAGAAAACGGGUGAGUAACAAUUGUUUAAACAUCACAUAACUAAUUGCGAAAAUAUGAAAACUAAAUCAGAAAAUA--U-U ((((((..((((((((((......))))))))))......))))))......((((((((............))).))))).............................--.-. ( -25.20) >DroEre_CAF1 185613 102 + 1 ACCCGAAUUGACCUUGGCGACCGAGUGAAGGUCACAGAAAACGGGUGAGUAGCAAAGGUUUGACCAU---GUAA-AAACUGUUAAGUUAUGA------AAUUA-AAAAUA--UUU (((((...(((((((.((......)).))))))).......)))))..(((((...(((((......---....-))))).....)))))..------.....-......--... ( -23.30) >DroAna_CAF1 163503 98 + 1 ACACGGCUUGACCUCGGCGACCGUGUCAAGGUCACGGAGAACUGGUAAGUAAAAACUGAACCAAUAUGUCAUAA-----------------AAAACUAAAACAUAAAAUAUAUUU ........(((((((.(.((((.......)))).).)))...((((.(((....)))..))))....))))...-----------------........................ ( -19.80) >consensus ACCCGUAUUGACCUUGGCGACCGAGUCAAGGUCACAGAAAACGGGUGAGUAACAAUUGUUUAAACAUCACAUAA_UAAUUGCGAAAAUAUGAAAACUAAAUCAGAAAAUA__U_U ((((((..((((((((((......))))))))))......))))))..................................................................... (-16.50 = -18.10 + 1.60)

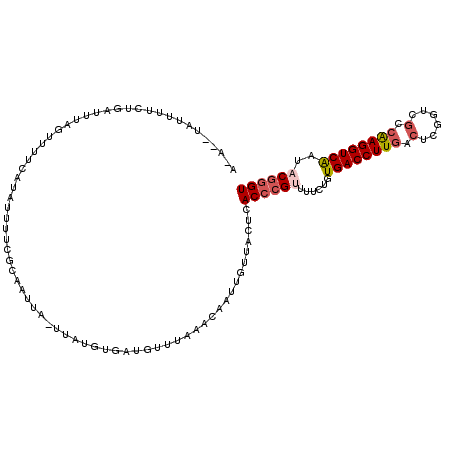
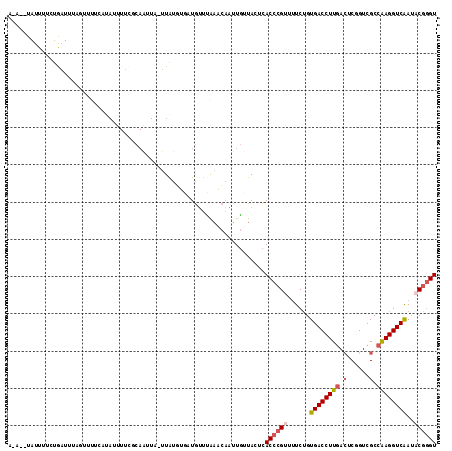
| Location | 2,042,496 – 2,042,608 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.16 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -15.04 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2042496 112 - 27905053 A-A--UAUUUUCCGAUUUUGUUUUAAUAUUUUCUCAAUUAUUUGUUUGCUGUUUAAACAAUUGUUACUCACCCGUUUUCUGUGACCUUGAAUCGGUCGCCAAGGUCGAUACGGGU .-.--........(((.((((((.((((......(((....))).....)))).))))))..)))....((((((......((((((((..........))))))))..)))))) ( -23.10) >DroSec_CAF1 185682 112 - 1 A-A--UAUUUUCUGAUUUAGUUUUCAUAUUUUCGCAAUUAGUUAUGUGCUGUUUAAACAAUUGUUACUCACCCGUUUUCUGUGACCUUGACUCUGUCGCCAAGGUCAAUACGGGU .-.--..............((((..(((....((((........)))).)))..))))...........((((((......((((((((.(......).))))))))..)))))) ( -23.10) >DroSim_CAF1 190586 112 - 1 A-A--UAUUUUCUGAUUUAGUUUUCAUAUUUUCGCAAUUAGUUAUGUGAUGUUUAAACAAUUGUUACUCACCCGUUUUCUGUGACCUUGACUCUGUCGCCAAGGUCAAUACGGGU .-.--..............((((..(((...(((((........))))))))..))))...........((((((......((((((((.(......).))))))))..)))))) ( -25.30) >DroEre_CAF1 185613 102 - 1 AAA--UAUUUU-UAAUU------UCAUAACUUAACAGUUU-UUAC---AUGGUCAAACCUUUGCUACUCACCCGUUUUCUGUGACCUUCACUCGGUCGCCAAGGUCAAUUCGGGU ...--......-.....------.................-....---..((.....))..........(((((...(((((((((.......))))))..)))......))))) ( -16.20) >DroAna_CAF1 163503 98 - 1 AAAUAUAUUUUAUGUUUUAGUUUU-----------------UUAUGACAUAUUGGUUCAGUUUUUACUUACCAGUUCUCCGUGACCUUGACACGGUCGCCGAGGUCAAGCCGUGU ........................-----------------...((((..((((((..(((....))).)))))).(((.((((((.......)))))).)))))))........ ( -22.90) >consensus A_A__UAUUUUCUGAUUUAGUUUUCAUAUUUUCGCAAUUA_UUAUGUGAUGUUUAAACAAUUGUUACUCACCCGUUUUCUGUGACCUUGACUCGGUCGCCAAGGUCAAUACGGGU .....................................................................((((((......((((((((.(......).))))))))..)))))) (-15.04 = -15.92 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:44 2006