
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,412,396 – 14,412,541 |
| Length | 145 |
| Max. P | 0.650355 |

| Location | 14,412,396 – 14,412,501 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -39.82 |
| Consensus MFE | -20.43 |
| Energy contribution | -24.13 |
| Covariance contribution | 3.70 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14412396 105 - 27905053 CCGCAGCUCCUGGUAAGGUUGCUGCA---CCAGCAACAUUAUUGCUAGACAGAUUGGCGGGUAUGGAGGUCAUGCUCCUGCUGCUUGCGGUUCCGUUUCCAGGUGCGA .....((.(((((.((.(..((((((---.(((((........(((((.....)))))(((((((.....))))))).)))))..))))))..).)).))))).)).. ( -41.20) >DroSec_CAF1 191018 102 - 1 GAGCAGCUCCUGGUAAGGUUGCUGCA---CCAGCAACAUUAUUGCUAGACAGAUUGGCGGGUAUGGAGGUCAUGCUCCU---GCUUGCGGUUCCGUUUCCAGCUGCGA ..((((((.(((((((.(((((((..---.)))))))....)))))))((.((.(.(((((((.((((......)))))---)))))).).)).))....)))))).. ( -43.10) >DroSim_CAF1 183113 102 - 1 GCGCAGCUCCUGGUAAGGUUGCUGCA---CCAGCAACAUUAUUGCUAGACAGAUUGGCGGGUAUGGAGGUCAUGCUCCU---GCUUUCGGUUCCGUUUCCAGCUGCGA .(((((((.(((((((.(((((((..---.)))))))....)))))))..........(((.((((((.(...((....---))....).)))))).)))))))))). ( -40.10) >DroEre_CAF1 198720 102 - 1 CCGCAGCUCCUGGCAAGGUUGCUGCU---CCAGCAACAUUAUUGCUAGACAGAUUGGCGGGAAUGGAGGUCAUGCUCCU---GCUUGCGGUUCCGUUUCCAGCUGCGA .(((((((.(((((((.(((((((..---.)))))))....)))))))..........((((((((((.(((.((....---)).)).).))))))))))))))))). ( -47.50) >DroWil_CAF1 227697 90 - 1 CCACACCACCGGCGGCAGCUGCUGCAGCAGCAGCCACAUUAUUGCUGGACAAAUUGGCAGGUAUGGAGGUCAUGCUUCG---GGCUG------CGU---------UGG .....(((.(.(((((....)))))....((((((.(((..((((..(.....)..))))..)))((((.....)))).---)))))------)).---------))) ( -33.40) >DroAna_CAF1 175625 96 - 1 CGGCUGCUCCGGCGAGAUUUGCUGCU---CCAGCAACGUUGUUGCUGGACAGAUUGGCGGGUAUGGAGGUCAUGCUCCU---GGUUCCAGUUCCGUUUUCAG------ ((((((.((((((((.(.((((((..---.))))))...).)))))))))))((..(.(((((((.....)))))))).---.)).......))).......------ ( -33.60) >consensus CCGCAGCUCCUGGUAAGGUUGCUGCA___CCAGCAACAUUAUUGCUAGACAGAUUGGCGGGUAUGGAGGUCAUGCUCCU___GCUUGCGGUUCCGUUUCCAGCUGCGA .((((.((.(((((((.(((((((......)))))))....)))))))((.((.(.((((((..((((......))))....)))))).).)).))....)).)))). (-20.43 = -24.13 + 3.70)

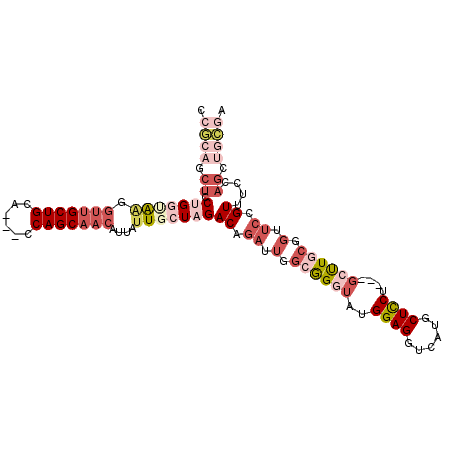

| Location | 14,412,436 – 14,412,541 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.77 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.71 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.39 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14412436 105 + 27905053 CUCCAUACCCGCCAAUCUGUCUAGCAAUAAUGUUGCUGG---UGCAGCAACCUUA---CCAGGAGCUGC---------GGCACCGCCAGAGAAAUAUACAUGGCCGAACUCCAAGGACGA .(((......(((.......((((((((...))))))))---.(((((..(((..---..))).)))))---------))).(.((((............)))).)........)))... ( -31.20) >DroVir_CAF1 232504 108 + 1 UUCCAUACCUGCCACGCUGUCCAGCAAUAAUGUGGCUGG---CACUGCG---GCGACCGCUGGCGCUGCCCUAG------UGGCGCCCGAGAAGCAGCCAUGGCCCAAUUCCAAGGAUGA ...(((.((((((..(((....)))......(((((((.---(...(((---(...)))).(((((..(....)------..)))))......))))))))))).........)))))). ( -39.60) >DroEre_CAF1 198757 105 + 1 CUCCAUUCCCGCCAAUCUGUCUAGCAAUAAUGUUGCUGG---AGCAGCAACCUUG---CCAGGAGCUGC---------GGCACCGCCAGAGAAACAGCCGUGGCCCAACUCCAAAGACGA .................(((((((((((...))))))((---((..(((....))---)..((.((..(---------(((...............))))..))))..))))..))))). ( -31.66) >DroWil_CAF1 227719 108 + 1 CUCCAUACCUGCCAAUUUGUCCAGCAAUAAUGUGGCUGCUGCUGCAGCAGCUGCC---GCCGGUGGUGU---------GGUCCCAGCAGAGAAGCAGCCGUGGCCCAACUCCAAGGAUGA ..(((((((.(((...(((.....)))....(((((.(((((....))))).)))---)).))))))))---------))(((..((......)).((....))..........)))... ( -38.30) >DroMoj_CAF1 239507 117 + 1 UUCCAUACCUGCCACGCUGUCCAGCAACAAUGUUGCAGC---AACAGCAGCUGCAACGGCUGCAGCUGGAUUGGGCGCCACGGCGCCAGAGAAACAGCCCUGGCCCAACUCCAAGGACGA .......(((((((.((((((((((..((.(((((((((---.......)))))))))..))..))))))...(((((....))))).......))))..)))).........))).... ( -48.50) >DroAna_CAF1 175656 105 + 1 CUCCAUACCCGCCAAUCUGUCCAGCAACAACGUUGCUGG---AGCAGCAAAUCUC---GCCGGAGCAGC---------CGCUCCGCCAGAGAAGCAGCCAUGGCCCAACUCCAAAGACGA ..........((((.....(((((((((...))))))))---)((.((...((((---..((((((...---------.))))))...)))).)).))..))))................ ( -37.70) >consensus CUCCAUACCCGCCAAUCUGUCCAGCAAUAAUGUUGCUGG___AGCAGCAACUGCA___GCAGGAGCUGC_________GGCACCGCCAGAGAAACAGCCAUGGCCCAACUCCAAGGACGA ..........((((..((((((((((((...))))))))....(((((................)))))........................))))...))))................ (-14.90 = -15.71 + 0.81)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:09 2006