
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,402,658 – 14,402,785 |
| Length | 127 |
| Max. P | 0.972991 |

| Location | 14,402,658 – 14,402,772 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.00 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -16.12 |
| Energy contribution | -15.40 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14402658 114 + 27905053 UGGUUAGAAAUUAGUCGACCUAUUUGGC------CAUUUUAAUAUUCCUGUACGCCAUUCGCUUAAUUGGUGGCAACAGGUGCCACACCUGCGCCUCUCUCUCGCCCCCCCCCCUUCUAC .(((.(((((((((.(((......((((------...................)))).))).))))))((.(((..((((((...)))))).))).)).))).))).............. ( -24.01) >DroSec_CAF1 181307 102 + 1 UGGAUUGAAAUUAGCCGACCUAUUUGGC------CAUUUUAAUAUUUCUAAACGCCAUUCGCUUAAUUGGUGGCAACAGGUGCCACACCUGCGUCUC-------CCC-----CCUUCUAC .((((((((((..(((((.....)))))------.))))))))..........((((((.........))))))..((((((...))))))......-------...-----))...... ( -22.70) >DroSim_CAF1 173367 107 + 1 UGGAUAGAAAUUAGCCGACCUAUUUGGC------CAUUUUAAUAUUUCUAAACGCCAUUCGCUUAAUUGGUGGCAACAGGUGCCACACCUGCGCCUC-------CCCCCUCCCCUUCUAC .((((((((((..(((((.....)))))------.........)))))))..................((.(((..((((((...)))))).))).)-------)....)))........ ( -24.70) >DroEre_CAF1 188586 95 + 1 UGGGUAAAAAUGUCCCAACCUAUC-GGC------CGUUUUAAUAUUUCUAAACGCCAUUCGCUUAAUUGGUGGCAACAGGUGCCACACCUGCGCC------------------CCGCCAC ((((.........)))).......-(((------.((((..........)))))))...........((((((...((((((...))))))....------------------)))))). ( -25.90) >DroYak_CAF1 202278 101 + 1 UGGGUAGAAACUAGCCAACCUAUU-GGUCGGUCCCAUUUUAAUAUUUCUAAACGCCAUUCGCUUAAUUGGUGGCAACAGGUGCCACACCUGCGCC------------------CUUCCAC .((((.(..((..(((((....))-)))..))..)..................((((((.........))))))..((((((...)))))).)))------------------)...... ( -26.20) >consensus UGGAUAGAAAUUAGCCGACCUAUUUGGC______CAUUUUAAUAUUUCUAAACGCCAUUCGCUUAAUUGGUGGCAACAGGUGCCACACCUGCGCCUC_______CCC_____CCUUCUAC (((...........)))........(((.........................((((((.........))))))..((((((...)))))).)))......................... (-16.12 = -15.40 + -0.72)

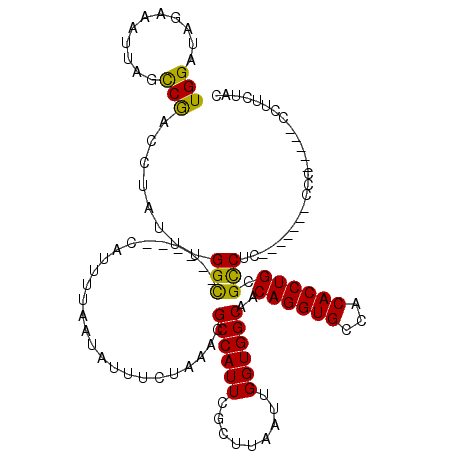

| Location | 14,402,658 – 14,402,772 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.00 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.710889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14402658 114 - 27905053 GUAGAAGGGGGGGGGGCGAGAGAGAGGCGCAGGUGUGGCACCUGUUGCCACCAAUUAAGCGAAUGGCGUACAGGAAUAUUAAAAUG------GCCAAAUAGGUCGACUAAUUUCUAACCA ......((..((..(...((.....((((((((((...)))))).)))).((......((.....)).....))..........((------(((.....))))).))..)..))..)). ( -31.20) >DroSec_CAF1 181307 102 - 1 GUAGAAGG-----GGG-------GAGACGCAGGUGUGGCACCUGUUGCCACCAAUUAAGCGAAUGGCGUUUAGAAAUAUUAAAAUG------GCCAAAUAGGUCGGCUAAUUUCAAUCCA ......((-----(..-------((((.(((((((...))))))).(((.....(((((((.....)))))))............(------(((.....)))))))...))))..))). ( -30.20) >DroSim_CAF1 173367 107 - 1 GUAGAAGGGGAGGGGG-------GAGGCGCAGGUGUGGCACCUGUUGCCACCAAUUAAGCGAAUGGCGUUUAGAAAUAUUAAAAUG------GCCAAAUAGGUCGGCUAAUUUCUAUCCA ......((..((..((-------(.((((((((((...)))))).)))).))..(((((((.....)))))))...........((------(((.........))))).)..))..)). ( -33.90) >DroEre_CAF1 188586 95 - 1 GUGGCGG------------------GGCGCAGGUGUGGCACCUGUUGCCACCAAUUAAGCGAAUGGCGUUUAGAAAUAUUAAAACG------GCC-GAUAGGUUGGGACAUUUUUACCCA .((((((------------------((((((((((...)))))).)))).))..(((((((.....))))))).............------)))-)......((((.........)))) ( -31.10) >DroYak_CAF1 202278 101 - 1 GUGGAAG------------------GGCGCAGGUGUGGCACCUGUUGCCACCAAUUAAGCGAAUGGCGUUUAGAAAUAUUAAAAUGGGACCGACC-AAUAGGUUGGCUAGUUUCUACCCA (((((((------------------((((((((((...)))))).)))).....(((((((.....))))))).............((.((((((-....))))))))..)))))))... ( -34.40) >consensus GUAGAAGG_____GGG_______GAGGCGCAGGUGUGGCACCUGUUGCCACCAAUUAAGCGAAUGGCGUUUAGAAAUAUUAAAAUG______GCCAAAUAGGUCGGCUAAUUUCUACCCA (((((((..................((((((((((...)))))).)))).((..(((((((.....)))))))...................(((.....))).))....)))))))... (-20.92 = -21.52 + 0.60)

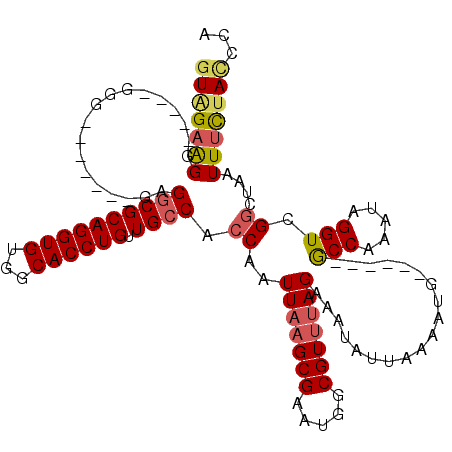

| Location | 14,402,692 – 14,402,785 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -17.52 |
| Consensus MFE | -14.89 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14402692 93 + 27905053 AAUAUUCCUGUACGCCAUUCGCUUAAUUGGUGGCAACAGGUGCCACACCUGCGCCUCUCUCUCGCCCCCCCCCCUUCUACCGCCAAAAGAAAA---- ....(((......((.....))....(((((((...((((((...)))))).((.........))..............)))))))..)))..---- ( -16.90) >DroSec_CAF1 181341 81 + 1 AAUAUUUCUAAACGCCAUUCGCUUAAUUGGUGGCAACAGGUGCCACACCUGCGUCUC-------CCC-----CCUUCUACCGCCAAAAGAAAA---- ....(((((....((.....))....(((((((...((((((...)))))).(....-------...-----.).....))))))).))))).---- ( -17.30) >DroSim_CAF1 173401 86 + 1 AAUAUUUCUAAACGCCAUUCGCUUAAUUGGUGGCAACAGGUGCCACACCUGCGCCUC-------CCCCCUCCCCUUCUACCGCCAAAAGAAAA---- ....(((((....((.....))......((.(((..((((((...)))))).))).)-------)......................))))).---- ( -17.90) >DroYak_CAF1 202317 79 + 1 AAUAUUUCUAAACGCCAUUCGCUUAAUUGGUGGCAACAGGUGCCACACCUGCGCC------------------CUUCCACCACGUAAAGAACCCCCA .....((((..((((.....)).....((((((.....(((((.......)))))------------------...)))))).))..))))...... ( -18.00) >consensus AAUAUUUCUAAACGCCAUUCGCUUAAUUGGUGGCAACAGGUGCCACACCUGCGCCUC_______CCC_____CCUUCUACCGCCAAAAGAAAA____ ....(((((....((((..........))))(((..((((((...)))))).)))................................)))))..... (-14.89 = -15.20 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:04 2006