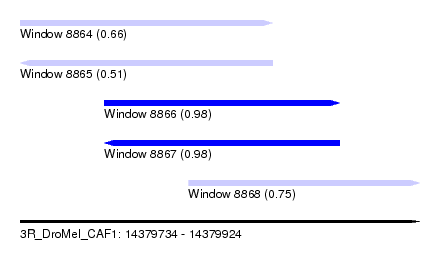
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,379,734 – 14,379,924 |
| Length | 190 |
| Max. P | 0.978399 |

| Location | 14,379,734 – 14,379,854 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.92 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14379734 120 + 27905053 AUUUCGUUUUUUUGGGUGGUGGACGAACGGGGGGCCAAUUAUGGCUAUAAAGUGUCUUCAUUAAAUUAUAAAUUCACGAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUAAAGUACU ...((((...((((..(((((((.((((....(((((....))))).....)).))))))))).....))))...))))((((...((((..(((((....)))))..)))).))))... ( -25.80) >DroPse_CAF1 170683 96 + 1 ------------------------GAACUGGGGGCCAAUUAUGGCAACAAAGUGUCUUCAUUAAAUUAUAAAUUCACGAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUGAAGUACU ------------------------....(((((((..(((.((....)).))))))))))...................((((..(((((..(((((....)))))..)))))))))... ( -20.80) >DroGri_CAF1 154741 93 + 1 ------------------G--------UUGU-GACCAAUUAUGGCUACAAAGUAUCUUCAUUAAAUUAUAAAUUCAUGAGCUUAACGUAAUUUGCAUGACAAUGCACUUUAUGAAGUACU ------------------.--------((((-(.(((....))).)))))((((.((((.((((.((((......))))..)))).((((..(((((....)))))..)))))))))))) ( -18.80) >DroWil_CAF1 188655 101 + 1 -------------------CGAACGACUGAGGGGCCAAUUAUGGCUACAAAGUGUCUUCAUUAAAUUAUAAAUUCACAAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUGAAGUACU -------------------.(((.(((.....(((((....))))).......))))))....................((((..(((((..(((((....)))))..)))))))))... ( -23.20) >DroAna_CAF1 145529 100 + 1 ACUUU-------UGGGU-------------GGGGCCAAUUAUGGCUACAAAGUGUCUUCAUUAAAUUAUAAAUUCACGAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUGAAGUACU (((((-------(((((-------------(..((((....))))..)...(((..((.((......)).))..)))..))))))(((((..(((((....)))))..)))))))))... ( -22.70) >DroPer_CAF1 175016 96 + 1 ------------------------GAACUGGGGGCCAAUUAUGGCAACAAAGUGUCUUCAUUAAAUUAUAAAUUCACGAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUGAAGUACU ------------------------....(((((((..(((.((....)).))))))))))...................((((..(((((..(((((....)))))..)))))))))... ( -20.80) >consensus ________________________GAACUGGGGGCCAAUUAUGGCUACAAAGUGUCUUCAUUAAAUUAUAAAUUCACGAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUGAAGUACU ..............................(..((((....))))..)...............................((((..(((((..(((((....)))))..)))))))))... (-15.75 = -16.00 + 0.25)

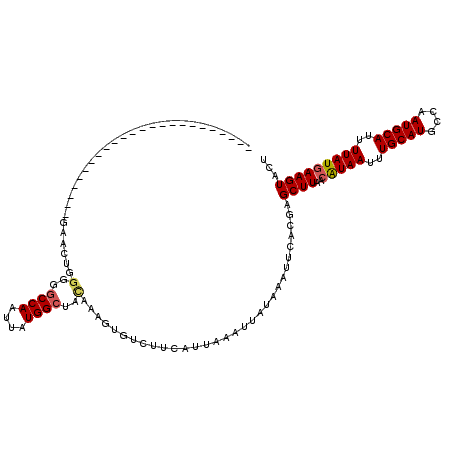

| Location | 14,379,734 – 14,379,854 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.92 |
| Mean single sequence MFE | -19.48 |
| Consensus MFE | -16.53 |
| Energy contribution | -16.45 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14379734 120 - 27905053 AGUACUUUAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUCGUGAAUUUAUAAUUUAAUGAAGACACUUUAUAGCCAUAAUUGGCCCCCCGUUCGUCCACCACCCAAAAAAACGAAAU .......((((((..((.((((((((.....))))))))))..(((..(((((......)))))..)))))))))((((....))))......(((((...............))))).. ( -19.66) >DroPse_CAF1 170683 96 - 1 AGUACUUCAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUCGUGAAUUUAUAAUUUAAUGAAGACACUUUGUUGCCAUAAUUGGCCCCCAGUUC------------------------ .((.((((((.....((.((((((((.....))))))))))...(((((.....)))))))))))))........((((....)))).........------------------------ ( -18.40) >DroGri_CAF1 154741 93 - 1 AGUACUUCAUAAAGUGCAUUGUCAUGCAAAUUACGUUAAGCUCAUGAAUUUAUAAUUUAAUGAAGAUACUUUGUAGCCAUAAUUGGUC-ACAA--------C------------------ ((((((((((.((((......(((((....(((...)))...))))).......)))).)))))).))))((((..(((....)))..-))))--------.------------------ ( -19.62) >DroWil_CAF1 188655 101 - 1 AGUACUUCAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUUGUGAAUUUAUAAUUUAAUGAAGACACUUUGUAGCCAUAAUUGGCCCCUCAGUCGUUCG------------------- .((.((((((.((((((.((((((((.....)))))))))).((((....)))))))).))))))))(((..(..((((....))))..)..)))......------------------- ( -20.90) >DroAna_CAF1 145529 100 - 1 AGUACUUCAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUCGUGAAUUUAUAAUUUAAUGAAGACACUUUGUAGCCAUAAUUGGCCCC-------------ACCCA-------AAAGU .((.((((((.....((.((((((((.....))))))))))...(((((.....)))))))))))))....((..((((....))))..)-------------)....-------..... ( -19.90) >DroPer_CAF1 175016 96 - 1 AGUACUUCAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUCGUGAAUUUAUAAUUUAAUGAAGACACUUUGUUGCCAUAAUUGGCCCCCAGUUC------------------------ .((.((((((.....((.((((((((.....))))))))))...(((((.....)))))))))))))........((((....)))).........------------------------ ( -18.40) >consensus AGUACUUCAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUCGUGAAUUUAUAAUUUAAUGAAGACACUUUGUAGCCAUAAUUGGCCCCCAGUUC________________________ .((.((((((.....((.((((((((.....))))))))))...(((((.....)))))))))))))........((((....))))................................. (-16.53 = -16.45 + -0.08)



| Location | 14,379,774 – 14,379,886 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.85 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -22.79 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14379774 112 + 27905053 AUGGCUAUAAAGUGUCUUCAUUAAAUUAUAAAUUCACGAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUAAAGUACUUUGUGGCCAUGAAAA------ACAUGAAAC--CGAUGAAC ((((((((((((((.(((..(((.....)))...............((((..(((((....)))))..)))).))))))))))))))))).....------.........--........ ( -24.90) >DroPse_CAF1 170699 114 + 1 AUGGCAACAAAGUGUCUUCAUUAAAUUAUAAAUUCACGAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUGAAGUACUUUGCGGCCAUGAAAA------ACACGAAAGCCCAAUGAAA (((((..(((((((.((((((.(((((((.................)))))))((((....)))).....)))))))))))))..))))).....------...(....).......... ( -26.93) >DroWil_CAF1 188676 109 + 1 AUGGCUACAAAGUGUCUUCAUUAAAUUAUAAAUUCACAAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUGAAGUACUUUGUGGCCAUGAAAA------AUACAA-----CGACGAAU ((((((((((((((.((((((.(((((((.................)))))))((((....)))).....)))))))))))))))))))).....------......-----........ ( -31.03) >DroMoj_CAF1 192285 117 + 1 AUGGCGACAAAGUGUCUUCAUUAAAUUAUAAAUUCAUGAGCUUAACGUAAUUUGCAUGCCAAUGCACUUUAUGAAGUACUUUGAG-CCAUUAAAAUGGAUAAAAAAAAAA--UAAGAAAA (((((..(((((((.((((.((((.((((......))))..)))).((((..(((((....)))))..))))))))))))))).)-))))....................--........ ( -25.50) >DroAna_CAF1 145549 112 + 1 AUGGCUACAAAGUGUCUUCAUUAAAUUAUAAAUUCACGAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUGAAGUACUUUGUGGCCAUGAAAC------ACAUGAAUC--CGAUGAAC ((((((((((((((.((((((.(((((((.................)))))))((((....)))).....)))))))))))))))))))).....------.........--........ ( -31.03) >DroPer_CAF1 175032 114 + 1 AUGGCAACAAAGUGUCUUCAUUAAAUUAUAAAUUCACGAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUGAAGUACUUUGCGGCCAUGAAAA------ACACGAAAGCUCCAUGAAA (((((..(((((((.((((((.(((((((.................)))))))((((....)))).....)))))))))))))..))))).....------...(....).......... ( -26.93) >consensus AUGGCUACAAAGUGUCUUCAUUAAAUUAUAAAUUCACGAGCUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUGAAGUACUUUGUGGCCAUGAAAA______ACACGAAAC__CGAUGAAA (((((..(((((((.((((((.(((((((.................)))))))((((....)))).....)))))))))))))..))))).............................. (-22.79 = -22.85 + 0.06)


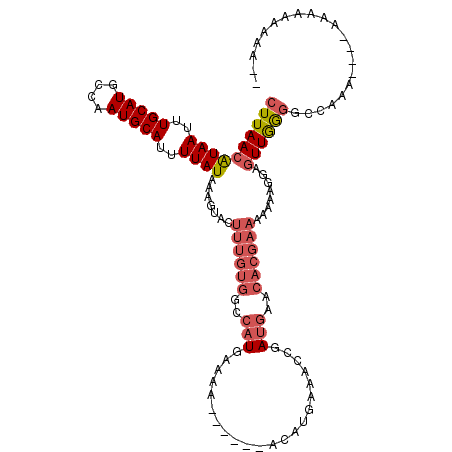
| Location | 14,379,774 – 14,379,886 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.85 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -23.70 |
| Energy contribution | -24.07 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14379774 112 - 27905053 GUUCAUCG--GUUUCAUGU------UUUUCAUGGCCACAAAGUACUUUAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUCGUGAAUUUAUAAUUUAAUGAAGACACUUUAUAGCCAU .......(--((..((((.------....)))))))..........(((((((..((.((((((((.....))))))))))..(((..(((((......)))))..)))))))))).... ( -18.70) >DroPse_CAF1 170699 114 - 1 UUUCAUUGGGCUUUCGUGU------UUUUCAUGGCCGCAAAGUACUUCAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUCGUGAAUUUAUAAUUUAAUGAAGACACUUUGUUGCCAU ...................------.....(((((.(((((((.((((((.....((.((((((((.....))))))))))...(((((.....)))))))))))..))))))).))))) ( -26.90) >DroWil_CAF1 188676 109 - 1 AUUCGUCG-----UUGUAU------UUUUCAUGGCCACAAAGUACUUCAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUUGUGAAUUUAUAAUUUAAUGAAGACACUUUGUAGCCAU ........-----......------.....(((((.(((((((.((((((.((((((.((((((((.....)))))))))).((((....)))))))).))))))..))))))).))))) ( -26.90) >DroMoj_CAF1 192285 117 - 1 UUUUCUUA--UUUUUUUUUUAUCCAUUUUAAUGG-CUCAAAGUACUUCAUAAAGUGCAUUGGCAUGCAAAUUACGUUAAGCUCAUGAAUUUAUAAUUUAAUGAAGACACUUUGUCGCCAU ........--....................((((-(.((((((.((((((.((((.(((.(((..((.......))...))).)))........)))).))))))..))))))..))))) ( -21.90) >DroAna_CAF1 145549 112 - 1 GUUCAUCG--GAUUCAUGU------GUUUCAUGGCCACAAAGUACUUCAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUCGUGAAUUUAUAAUUUAAUGAAGACACUUUGUAGCCAU ........--((..(....------)..))(((((.(((((((.((((((.....((.((((((((.....))))))))))...(((((.....)))))))))))..))))))).))))) ( -26.90) >DroPer_CAF1 175032 114 - 1 UUUCAUGGAGCUUUCGUGU------UUUUCAUGGCCGCAAAGUACUUCAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUCGUGAAUUUAUAAUUUAAUGAAGACACUUUGUUGCCAU ...((((((...)))))).------.....(((((.(((((((.((((((.....((.((((((((.....))))))))))...(((((.....)))))))))))..))))))).))))) ( -29.10) >consensus UUUCAUCG__CUUUCGUGU______UUUUCAUGGCCACAAAGUACUUCAUAAAAUGCAUUGGCAUGCAAAUUAUGUUAAGCUCGUGAAUUUAUAAUUUAAUGAAGACACUUUGUAGCCAU ..............................(((((.(((((((.((((((.....((.((((((((.....))))))))))...(((((.....)))))))))))..))))))).))))) (-23.70 = -24.07 + 0.36)



| Location | 14,379,814 – 14,379,924 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -19.49 |
| Consensus MFE | -13.25 |
| Energy contribution | -14.50 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14379814 110 + 27905053 CUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUAAAGUACUUUGUGGCCAUGAAAA------ACAUGAAACCGAUGAACACGAAAAAAAGGAGUUGGGGCAAAAAAAAAAAAAAAGA-- ((((((((((..(((((....)))))..)))).......((((((..(((.....------...........)))..))))))........))))))...................-- ( -18.99) >DroSec_CAF1 158592 105 + 1 CUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUAAAGUACUUUGUGGCCAUGAAAA------ACAUGAAACCGAUGAACACGAAAAAAAGGAGUUGGGGACAAA----AAA-AAAGA-- ((((((((((..(((((....)))))..)))).......((((((..(((.....------...........)))..))))))........))))))......----...-.....-- ( -18.99) >DroSim_CAF1 153096 106 + 1 CUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUAAAGUACUUUGUGGCCAUGAAAA------ACAUGAAACCGAUGAACACGAAAAAAAGGAGUUGGGGACAAA----AAAAAAAGA-- ((((((((((..(((((....)))))..)))).......((((((..(((.....------...........)))..))))))........))))))......----.........-- ( -18.99) >DroEre_CAF1 166004 103 + 1 CUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUAAAGUACUUUGUGGCCAUGAAAA------ACAUGAAACCGAUGAACACGAAA--AAGGAGUUGGGGCCCAA----A-AAAAUAA-- ......((((..(((((....)))))..)))).......((((.(((((((....------.)))....(((((...(......--..)..))))))))))))----)-.......-- ( -22.10) >DroYak_CAF1 178375 105 + 1 CUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUAAAGUACUUUGUGGCCAUGAAAA------ACAUGAAACCGAUGAACACGAAA-AAAGGAGUUGGGGCCCAA----AUAAAAAAA-- ......((((..(((((....)))))..)))).......((((.(((((((....------.)))....(((((...(......-...)..))))))))))))----)........-- ( -21.10) >DroMoj_CAF1 192325 107 + 1 CUUAACGUAAUUUGCAUGCCAAUGCACUUUAUGAAGUACUUUGAG-CCAUUAAAAUGGAUAAAAAAAAAAUAAGAAAAAA--CCUC----AUUUAAAUCGCAA----ACAAAAUACAU .....(((((..(((((....)))))..)))))..(((.((((.(-(.(((.((((((......................--..))----)))).))).))..----.)))).))).. ( -16.76) >consensus CUUAACAUAAUUUGCAUGCCAAUGCAUUUUAUAAAGUACUUUGUGGCCAUGAAAA______ACAUGAAACCGAUGAACACGAAAAAAAGGAGUUGGGGCCAAA____AAAAAAAAA__ ((((((((((..(((((....)))))..)))).......((((((..(((......................)))..))))))........))))))..................... (-13.25 = -14.50 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:58 2006