| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,372,504 – 14,372,608 |
| Length | 104 |
| Max. P | 0.997272 |

| Location | 14,372,504 – 14,372,608 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 69.66 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.13 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994543 |
| Prediction | RNA |
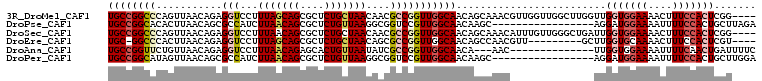
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14372504 104 + 27905053 ----CCGAGUGGAAAGUUUUCCACCAACCAAGCCAACCAACGUUUGCUGUUGCCAACCGGCGUUGUUAGCAGAGCGCUGCUAAAGGACCUCUGUUAACUGGGCCGGCA ----....((((((....))))))((((..(((.(((....))).)))))))....(((((.(.((((((((((.(((......)).))))))))))).).))))).. ( -38.00) >DroPse_CAF1 161812 91 + 1 UCUAAGCAGUGGAAAAUUUUCCAUCCU-----------------GCUUGUUGCCAACGGACCGCCUUAACAGAGCGCUGUUAAGAUGGCGCUGUUAAGUGUGCCGGCA ..((((((((((((....)))))..))-----------------))))).((((..........((((((((....))))))))..(((((........))))))))) ( -35.20) >DroSec_CAF1 151214 104 + 1 ----CCGAGUGGAAAGUUUUCCACCAAUCAGCCCAACAAAUGUUUGCUGUUGCCAACCGGCGUUGUUAGCAGAGCGCUGUUAAAGGACCUCUGUUAACUGGGCCGGCA ----....((((((....))))))(((.((((..(((....))).)))))))....(((((.(.((((((((((.(((......)).))))))))))).).))))).. ( -38.60) >DroEre_CAF1 158456 94 + 1 ----ACGAGUGGAAAGUUUUGCACCAAGC---------AACGUUGGCUGUUGCCAACCGGCGCUGUUAGCAGAGCGCUGCUAAAGGACCUCUGUUAAGUGGGCC-GCA ----..(.(((.((....)).))))..((---------...((((((....)))))).(((.(..(((((((((.(((......)).))))))))))..).)))-)). ( -34.00) >DroAna_CAF1 139492 91 + 1 GAAAAUCAGUUGAAAAUUUUCCACCAA--------------GUU---UGUUGCCAACCGGCGAUAUUAACAGUGCUCUGUUAAAGGACCUCUGUUAACAGAACCGGCA ........((((.......(((.....--------------...---(((((((....)))))))(((((((....))))))).)))..((((....))))..)))). ( -23.60) >DroPer_CAF1 166290 91 + 1 UCCAAGCAGUGGAAAAUUUUCCAUCCU-----------------GCUUGUUGCCAACGGACCGCCUUAACAGAGCGCUGUUAAGAUGGCGCUGUUAACUAUGCCGGCA ..((((((((((((....)))))..))-----------------))))).((((..........((((((((....))))))))..((((..(....)..)))))))) ( -33.50) >consensus ____ACCAGUGGAAAAUUUUCCACCAA______________GUUGGCUGUUGCCAACCGGCGGUGUUAACAGAGCGCUGUUAAAGGACCUCUGUUAACUGGGCCGGCA ........((((((....))))))..........................((((...........(((((((....))))))).....(((........)))..)))) (-15.52 = -15.13 + -0.39)

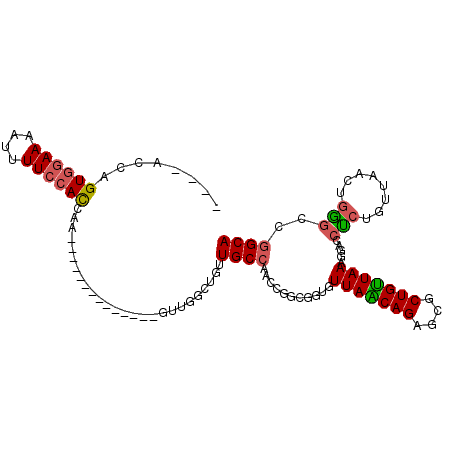
| Location | 14,372,504 – 14,372,608 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.66 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -17.53 |
| Energy contribution | -18.42 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14372504 104 - 27905053 UGCCGGCCCAGUUAACAGAGGUCCUUUAGCAGCGCUCUGCUAACAACGCCGGUUGGCAACAGCAAACGUUGGUUGGCUUGGUUGGUGGAAAACUUUCCACUCGG---- .(((((((.(((((((.........(((((((....)))))))(((((...((((....))))...)))))))))))).)))))))((((....))))......---- ( -42.10) >DroPse_CAF1 161812 91 - 1 UGCCGGCACACUUAACAGCGCCAUCUUAACAGCGCUCUGUUAAGGCGGUCCGUUGGCAACAAGC-----------------AGGAUGGAAAAUUUUCCACUGCUUAGA (((((((((.(((((((((((..........))))..)))))))...))..)))))))..((((-----------------((..(((((....)))))))))))... ( -33.00) >DroSec_CAF1 151214 104 - 1 UGCCGGCCCAGUUAACAGAGGUCCUUUAACAGCGCUCUGCUAACAACGCCGGUUGGCAACAGCAAACAUUUGUUGGGCUGAUUGGUGGAAAACUUUCCACUCGG---- ...(((((((((((.(((((((.........)).))))).)))).......((((....))))..........)))))))...(((((((....)))))))...---- ( -36.30) >DroEre_CAF1 158456 94 - 1 UGC-GGCCCACUUAACAGAGGUCCUUUAGCAGCGCUCUGCUAACAGCGCCGGUUGGCAACAGCCAACGUU---------GCUUGGUGCAAAACUUUCCACUCGU---- ...-.(((((..((((...(((.(((((((((....))))))).)).)))(((((....)))))...)))---------)..))).))................---- ( -31.70) >DroAna_CAF1 139492 91 - 1 UGCCGGUUCUGUUAACAGAGGUCCUUUAACAGAGCACUGUUAAUAUCGCCGGUUGGCAACA---AAC--------------UUGGUGGAAAAUUUUCAACUGAUUUUC .((((((..(((((((((.(.((........)).).)))))))))..))))))((....))---...--------------(..((.(((....))).))..)..... ( -26.30) >DroPer_CAF1 166290 91 - 1 UGCCGGCAUAGUUAACAGCGCCAUCUUAACAGCGCUCUGUUAAGGCGGUCCGUUGGCAACAAGC-----------------AGGAUGGAAAAUUUUCCACUGCUUGGA (((((((...((.....))(((..((((((((....))))))))..)))..))))))).(((((-----------------((..(((((....)))))))))))).. ( -37.70) >consensus UGCCGGCCCAGUUAACAGAGGUCCUUUAACAGCGCUCUGCUAACAACGCCGGUUGGCAACAGCCAAC______________UUGGUGGAAAACUUUCCACUCGU____ ((((((((...........(((...(((((((....)))))))....))))))))))).........................(((((((....)))))))....... (-17.53 = -18.42 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:53 2006