| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,367,527 – 14,367,629 |
| Length | 102 |
| Max. P | 0.952425 |

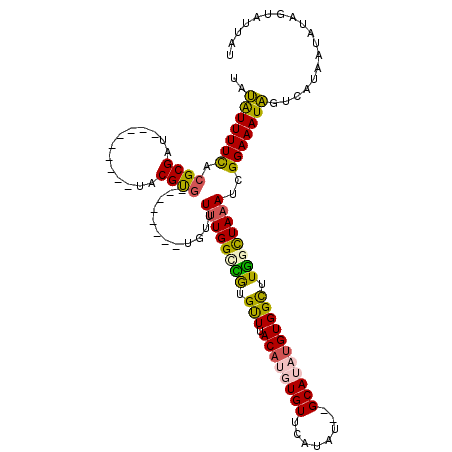
| Location | 14,367,527 – 14,367,629 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -18.23 |
| Energy contribution | -18.14 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14367527 102 - 27905053 UAUAUUUUCACGCGAU---------UACGUG--------UGUUUUUGGCCGUGUUUACAUGUGUUCAUAUGUGCAUAUGUGGCUUGGCUAAAUCGGAAAUAGUCAUAAUAUAGUAUUAU ..((((((((((((..---------..))))--------)...((((((((.(((.((((((((........))))))))))).))))))))..))))))).................. ( -29.50) >DroSec_CAF1 146235 102 - 1 UAUAUUUUCACGCGAU---------UACGUG--------UGCUUUUGGCCGUGUUUACAUGUGUUCAUAUGUGCAUAUGUGGCUUGGCUAAAUCGGAAAUAGUCAUAAUAUAGUAUUAU ..((((((((((((..---------..))))--------)...((((((((.(((.((((((((........))))))))))).))))))))..))))))).................. ( -29.50) >DroSim_CAF1 140595 102 - 1 UAUAUUUUCACGCGAU---------UACGUG--------UGUUUUUGGCCGUGUUUACAUGUGUUCAUAUGUGCAUAUGUGGCUUGGCUAAAUCGGAAAUAGUCAUAAUAUAGUAUUAU ..((((((((((((..---------..))))--------)...((((((((.(((.((((((((........))))))))))).))))))))..))))))).................. ( -29.50) >DroEre_CAF1 153713 108 - 1 -AUAUUUUCACGCGAUUCGCACGAUUACGUG--------UGUUUUUGGUUGUGCUUACAUGUGUUCAUAC--GCAAUUGUGGCCUGGCUAAAUUGGAAAUAUUCAUAAUAUGGCAUUAU -(((((((((.......((((((....))))--------))..((((((((.(((.((((((((....))--)))..)))))).)))))))).)))))))))................. ( -25.60) >DroYak_CAF1 155191 107 - 1 -AAAUUUUCACGCGAU---------UACGUGUGUUUUUGUGUUUUUGGGUGUGUUUACAUGUGUUCAUAU--GCAUAUGUGGCUUGGCUAAAUUGGAAAUUUUCAUAAAAUAGUAUUAU -.......((((((..---------..)))))).(((((((.((((.(((..((((((((((((......--)))))))))....)))...))).))))....)))))))......... ( -23.10) >DroAna_CAF1 134943 86 - 1 UCUGUUUUUCGGCGAU---------UACGCG--------UGUUUGUGGGUAUGUUUACU----------------AAUGUGGUUUAGUUAAAACGGAAAUAGUCAUAAUAUGGAAUUAU ((((((((..(((...---------..((((--------(....(((((....))))).----------------.))))).....)))))))))))...................... ( -13.80) >consensus UAUAUUUUCACGCGAU_________UACGUG________UGUUUUUGGCCGUGUUUACAUGUGUUCAUAU__GCAUAUGUGGCUUGGCUAAAUCGGAAAUAGUCAUAAUAUAGUAUUAU ..(((((((.((((.............))))............((((((((.(((.((((((((........))))))))))).))))))))..))))))).................. (-18.23 = -18.14 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:50 2006