
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,353,408 – 14,353,571 |
| Length | 163 |
| Max. P | 0.994393 |

| Location | 14,353,408 – 14,353,498 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 83.83 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -21.07 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14353408 90 + 27905053 UUGUUAUAUGCACAAAUGUGUCAAGCAUUACAGCAGCAGUAGCAGAAGCAGGCAUCUGCAUCUGCUGUCAGUGGUGGCAUCUCGUGGAUA ......(((.(((....((((((..((((...(((((((..(((((.(....).)))))..))))))).)))).))))))...))).))) ( -30.60) >DroSec_CAF1 132187 84 + 1 UUGUUAUAUGCACAAAUGUGUCAAGCAUUACAGCAGCAGCAGCAGCAGC------CUGCAUCUGCUGUCAGUGAUGGCAUCUCGUGGAUA ......(((.(((....((((((..(((((((((((..(((((....).------))))..))))))).)))).))))))...))).))) ( -28.20) >DroSim_CAF1 127870 89 + 1 UUGUUAUAUGCACAAAUGUGUCAAGCAUUACAGCAGCAGCAGCAGCAUCU-GCAUCUGCAUCUGCUGUCAGUGGUGGCAUCUCGUGGAUA ......(((.(((....((((((..(((((((((((..((((..((....-))..))))..))))))).)))).))))))...))).))) ( -31.30) >DroYak_CAF1 141176 68 + 1 UUGUUAUAUGCACAAAUGUGUCAAGCAUUACAGCAGCAG-------------------CAUCUGCUGUCAGU---GGCACCUCGUAGAUA .................((((((....(.(((((((...-------------------...))))))).).)---))))).......... ( -16.10) >consensus UUGUUAUAUGCACAAAUGUGUCAAGCAUUACAGCAGCAGCAGCAGCAGC______CUGCAUCUGCUGUCAGUGGUGGCAUCUCGUGGAUA ......(((.(((....((((((..(((((((((((..((((.............))))..))))))).)))).))))))...))).))) (-21.07 = -21.70 + 0.63)



| Location | 14,353,408 – 14,353,498 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 83.83 |
| Mean single sequence MFE | -22.25 |
| Consensus MFE | -16.39 |
| Energy contribution | -16.45 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14353408 90 - 27905053 UAUCCACGAGAUGCCACCACUGACAGCAGAUGCAGAUGCCUGCUUCUGCUACUGCUGCUGUAAUGCUUGACACAUUUGUGCAUAUAACAA (((.((((((.((.((......(((((((..(((((.(....).))))).....)))))))......)).))..)))))).)))...... ( -24.00) >DroSec_CAF1 132187 84 - 1 UAUCCACGAGAUGCCAUCACUGACAGCAGAUGCAG------GCUGCUGCUGCUGCUGCUGUAAUGCUUGACACAUUUGUGCAUAUAACAA (((.((((((.((.((......(((((((..((((------.(....)))))..)))))))......)).))..)))))).)))...... ( -23.10) >DroSim_CAF1 127870 89 - 1 UAUCCACGAGAUGCCACCACUGACAGCAGAUGCAGAUGC-AGAUGCUGCUGCUGCUGCUGUAAUGCUUGACACAUUUGUGCAUAUAACAA (((.((((((.((.((......(((((((..((((..((-....))..))))..)))))))......)).))..)))))).)))...... ( -24.60) >DroYak_CAF1 141176 68 - 1 UAUCUACGAGGUGCC---ACUGACAGCAGAUG-------------------CUGCUGCUGUAAUGCUUGACACAUUUGUGCAUAUAACAA (((.(((((((((.(---(...(((((((...-------------------...)))))))......)).))).)))))).)))...... ( -17.30) >consensus UAUCCACGAGAUGCCACCACUGACAGCAGAUGCAG______GCUGCUGCUGCUGCUGCUGUAAUGCUUGACACAUUUGUGCAUAUAACAA (((.(((.(((((.........(((((((..((((..........)))).....)))))))...........)))))))).)))...... (-16.39 = -16.45 + 0.06)


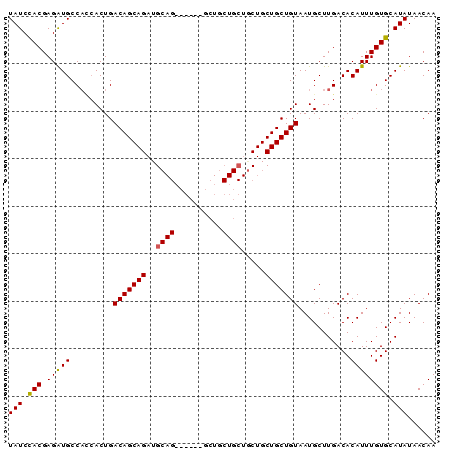
| Location | 14,353,432 – 14,353,538 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -39.97 |
| Consensus MFE | -38.79 |
| Energy contribution | -38.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14353432 106 + 27905053 GCAUUACAGCAGCAGUAGCAGAAGCAGGCAUCUGCAUCUGCUGUCAGUGGUGGCAUCUCGUGGAUAUAUAUCUCGAGGAUAUAUAUCUCGGGGAUAUAUAUCUUUU .((((((.(((((((..(((((.(....).)))))..)))))))..)))))).........(((((((((((((..((((....))))..)))))))))))))... ( -42.10) >DroSec_CAF1 132211 99 + 1 GCAUUACAGCAGCAGCAGCAGCAGC------CUGCAUCUGCUGUCAGUGAUGGCAUCUCGUGGAUAUAUAUCUCGAGGAUAUAUAUCUCGGGGAUAUAUAUCUU-U .((((((.(((((((..((((....------))))..)))))))..)))))).........(((((((((((((..((((....))))..))))))))))))).-. ( -39.30) >DroSim_CAF1 127894 104 + 1 GCAUUACAGCAGCAGCAGCAGCAUCU-GCAUCUGCAUCUGCUGUCAGUGGUGGCAUCUCGUGGAUAUAUAUCUCGAGGAUAUAUAUCUCGGGGAUAUAUAUCUU-U .(((((((((((..((((..((....-))..))))..))))))).))))............(((((((((((((..((((....))))..))))))))))))).-. ( -38.50) >consensus GCAUUACAGCAGCAGCAGCAGCAGC__GCAUCUGCAUCUGCUGUCAGUGGUGGCAUCUCGUGGAUAUAUAUCUCGAGGAUAUAUAUCUCGGGGAUAUAUAUCUU_U .((((((.(((((((..((((..........))))..)))))))..)))))).........(((((((((((((..((((....))))..)))))))))))))... (-38.79 = -38.57 + -0.22)



| Location | 14,353,432 – 14,353,538 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -29.33 |
| Energy contribution | -29.33 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14353432 106 - 27905053 AAAAGAUAUAUAUCCCCGAGAUAUAUAUCCUCGAGAUAUAUAUCCACGAGAUGCCACCACUGACAGCAGAUGCAGAUGCCUGCUUCUGCUACUGCUGCUGUAAUGC ....((((((((((..((((.........)))).))))))))))..................(((((((..(((((.(....).))))).....)))))))..... ( -32.80) >DroSec_CAF1 132211 99 - 1 A-AAGAUAUAUAUCCCCGAGAUAUAUAUCCUCGAGAUAUAUAUCCACGAGAUGCCAUCACUGACAGCAGAUGCAG------GCUGCUGCUGCUGCUGCUGUAAUGC .-..((((((((((..((((.........)))).))))))))))..........(((.((.(.((((((..((((------....))))..))))))).)).))). ( -31.80) >DroSim_CAF1 127894 104 - 1 A-AAGAUAUAUAUCCCCGAGAUAUAUAUCCUCGAGAUAUAUAUCCACGAGAUGCCACCACUGACAGCAGAUGCAGAUGC-AGAUGCUGCUGCUGCUGCUGUAAUGC .-..((((((((((..((((.........)))).))))))))))..................(((((((..((((..((-....))..))))..)))))))..... ( -33.40) >consensus A_AAGAUAUAUAUCCCCGAGAUAUAUAUCCUCGAGAUAUAUAUCCACGAGAUGCCACCACUGACAGCAGAUGCAGAUGC__GCUGCUGCUGCUGCUGCUGUAAUGC ....((((((((((..((((.........)))).))))))))))..................(((((((..((((..........)))).....)))))))..... (-29.33 = -29.33 + -0.00)


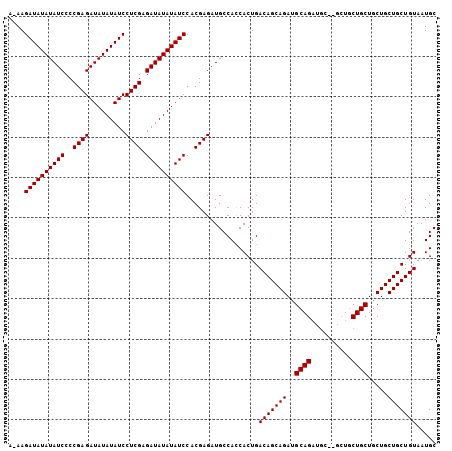
| Location | 14,353,468 – 14,353,571 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -22.35 |
| Energy contribution | -24.10 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14353468 103 + 27905053 UCUGCUGUCAGUGGUGGCAUCUCGUGGAUAUAUAUCUCGAGGAUAUAUAUCUCGGGGAUAUAUAUCUUUUUCGGGUGCACACACAAAUGUACAUCGCCUGCAG ....((((..(((((((((((.((.(((((((((((((..((((....))))..)))))))))))))....)))))))(((......))).))))))..)))) ( -38.30) >DroSec_CAF1 132241 102 + 1 UCUGCUGUCAGUGAUGGCAUCUCGUGGAUAUAUAUCUCGAGGAUAUAUAUCUCGGGGAUAUAUAUCUU-UUCGGGUGCACACACAAAUGUACAACCCCUUUAG ..........(((.(((((((.((.(((((((((((((..((((....))))..))))))))))))).-..))))))).)))))................... ( -32.50) >DroSim_CAF1 127929 102 + 1 UCUGCUGUCAGUGGUGGCAUCUCGUGGAUAUAUAUCUCGAGGAUAUAUAUCUCGGGGAUAUAUAUCUU-UUCGGGUGCACACACAAAUGUACAACCCCUUCAG .....(((((.((((((((((.((.(((((((((((((..((((....))))..))))))))))))).-..))))))).))).))..)).))).......... ( -33.90) >DroYak_CAF1 141217 81 + 1 UCUGCUGUCAGU---GGCACCUCGUAGAUAUAUA---------------UCUCGGGGAUAUGUAUCUU-UU-GGGUCCAUAC--AAAAAUACAUCCCCUUCAG .....(((..((---((.((((...(((((((((---------------((.....))))))))))).-..-))))))))))--).................. ( -22.60) >consensus UCUGCUGUCAGUGGUGGCAUCUCGUGGAUAUAUAUCUCGAGGAUAUAUAUCUCGGGGAUAUAUAUCUU_UUCGGGUGCACACACAAAUGUACAACCCCUUCAG ..........(((...((((((...(((((((((((((..((((....))))..))))))))))))).....)))))).)))..................... (-22.35 = -24.10 + 1.75)



| Location | 14,353,468 – 14,353,571 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -17.98 |
| Energy contribution | -20.10 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14353468 103 - 27905053 CUGCAGGCGAUGUACAUUUGUGUGUGCACCCGAAAAAGAUAUAUAUCCCCGAGAUAUAUAUCCUCGAGAUAUAUAUCCACGAGAUGCCACCACUGACAGCAGA ((((.((.(.(((((((....))))))))))......((((((((((..((((.........)))).)))))))))).....................)))). ( -30.00) >DroSec_CAF1 132241 102 - 1 CUAAAGGGGUUGUACAUUUGUGUGUGCACCCGAA-AAGAUAUAUAUCCCCGAGAUAUAUAUCCUCGAGAUAUAUAUCCACGAGAUGCCAUCACUGACAGCAGA ......((((.((((((....))))))))))((.-..((((((((((.....))))))))))((((.(((....)))..))))......)).(((....))). ( -29.20) >DroSim_CAF1 127929 102 - 1 CUGAAGGGGUUGUACAUUUGUGUGUGCACCCGAA-AAGAUAUAUAUCCCCGAGAUAUAUAUCCUCGAGAUAUAUAUCCACGAGAUGCCACCACUGACAGCAGA (((.((((((.((((((....))))))))))...-..((((((((((..((((.........)))).))))))))))...............))..))).... ( -29.70) >DroYak_CAF1 141217 81 - 1 CUGAAGGGGAUGUAUUUUU--GUAUGGACCC-AA-AAGAUACAUAUCCCCGAGA---------------UAUAUAUCUACGAGGUGCC---ACUGACAGCAGA (((..((((((((((((((--...((....)-))-))))))))..))))).(((---------------(....))))..........---........))). ( -19.10) >consensus CUGAAGGGGAUGUACAUUUGUGUGUGCACCCGAA_AAGAUAUAUAUCCCCGAGAUAUAUAUCCUCGAGAUAUAUAUCCACGAGAUGCCACCACUGACAGCAGA ......(((.(((((((....))))))))))......((((((((((..((((.........)))).))))))))))...............(((....))). (-17.98 = -20.10 + 2.12)

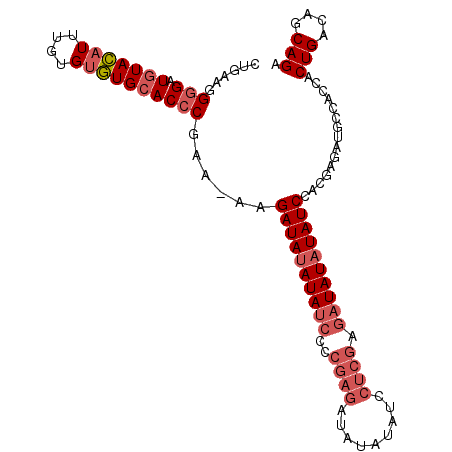

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:45 2006