| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,340,947 – 14,341,065 |
| Length | 118 |
| Max. P | 0.787057 |

| Location | 14,340,947 – 14,341,065 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.71 |
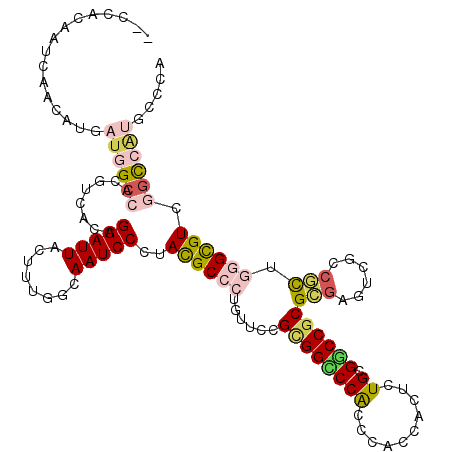
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -24.82 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.787057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
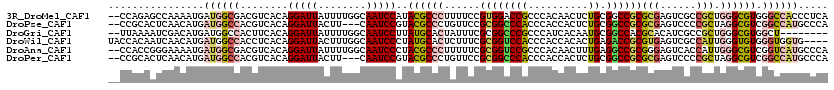
>3R_DroMel_CAF1 14340947 118 - 27905053 --CCAGAGCCAAAAUGAUGGCGACGUCACAGGAUUAUUUUGGCAAUCCAUACGCCCUUUUCCGUGGACCGCCCACAACUCUGCGGCCGCGCGAGUCGCCGCUGGGCGUGGGCCACCCUCA --...(.(((....(((((....)))))..(((((........))))).(((((((......((((.....))))......(((((.((....)).))))).)))))))))))....... ( -47.50) >DroPse_CAF1 128006 115 - 1 --CCGCACUCAACAUGAUGGCCACGUCACAGGAUUACUU---CAAUCCGUACGCCCUGUUCCGCGGCCCACCCACCACUCUGCGGCCGCGCGAGUCCCCGCUAGGCGUCGGCCAUGCCCA --..............((((((........(((((....---.)))))(.(((((.......((((((((..........)).))))))(((......)))..))))))))))))..... ( -40.30) >DroGri_CAF1 122395 110 - 1 --UUAAAAUCGACAUGAUGGCCACUUCACAGGAUUAUUUUGGCAAUCCUUAUGCACUAUUUCGCGGCCCGCCCAUCACAAUGCGGCCACGCACAUCGCCGCUGGGCGUGGCU-------- --.......(((((.(((((((........)).))))).))(((.......)))......))).((((((((((.......(((((..........))))))))))).))))-------- ( -30.91) >DroWil_CAF1 139845 116 - 1 UACCACAAUCAACAUGAUGGCCACCUCACAGGAUUACUUUGGCAAUCCCUAUGCACUCUUUCGCGGUCCACCCACCACACUGAGACCGCGUGAGUCGCCAUUGGGUGUGGGUGGUG---- ((((((.(((.....)))..((((((((..(((((.(....).)))))....((((((...(((((((((..........)).))))))).)))).))...)))).))))))))))---- ( -40.60) >DroAna_CAF1 112760 118 - 1 --CCACCGGGAAAAUGAUGGCGACGUCACAGGAUUAUUUUGGCAAUCCCUACGCCCUUUUUCGCGGUCCGCCCACAACUUUGAGGCCGCGGGAGUCACCAUUGGGCGUCGGUCAUGCCCA --.....(((...(((((..((((......(((((........)))))....((((.((..(((((((((..........)).)))))))..))........))))))))))))).))). ( -40.40) >DroPer_CAF1 130635 115 - 1 --CCGCACUCAACAUGAUGGCCACGUCACAGGAUUACUU---CAAUCCGUACGCCCUGUUCCGCGGCCCACCCACCACUCUGCGGCCGCGCGAGUCCCCGCUAGGCGUCGGCCAUGCCCA --..............((((((........(((((....---.)))))(.(((((.......((((((((..........)).))))))(((......)))..))))))))))))..... ( -40.30) >consensus __CCACAAUCAACAUGAUGGCCACGUCACAGGAUUACUUUGGCAAUCCCUACGCCCUGUUCCGCGGCCCACCCACCACUCUGCGGCCGCGCGAGUCGCCGCUGGGCGUCGGCCAUGCCCA ................((((((........(((((........)))))..((((((......((((((((..........)).))))))(((......))).)))))).))))))..... (-24.82 = -25.68 + 0.87)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:37 2006