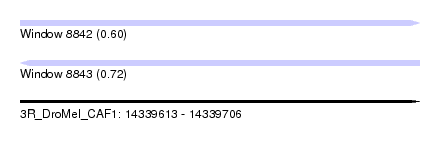
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,339,613 – 14,339,706 |
| Length | 93 |
| Max. P | 0.715351 |

| Location | 14,339,613 – 14,339,706 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -16.16 |
| Energy contribution | -15.52 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14339613 93 + 27905053 UA-AUUUCUGCAUUUUACAUGGGUGCAUAUC---UCGCCGUGGUGCUCG-GUUUAUGCCUA-GGACUUAGGGCUAC-UAGUAGUGUUUAGGCAUAAACU--------A------ ..-.....(((((((.....)))))))....---.(((....)))...(-(((((((((((-((((((((.....)-))..))).))))))))))))))--------.------ ( -27.60) >DroVir_CAF1 143373 94 + 1 AA-AAUUUUGCAUUUUACAAU-GUGCU--------CAUU-AGGUGCGCGUGUCUAUGCCUA-GGACUUAGGAUUGACUAGUAGUUGUUAGGCAUAAACU--------UGAAUCG ..-................((-((((.--------(...-..).))))))((.((((((((-((((((((......)))..)))).))))))))).)).--------....... ( -22.50) >DroPse_CAF1 126530 91 + 1 UAAAUUUUUGCAUUUUACAUG-GUGCA--UC---UCGCC-UGGUGCUCG-GUCUAUGCCUAUGGGCUUAGGACUAC-UAAUAGUGAUUAGGCAUAAACU--------A------ ........((((((......)-)))))--..---..(((-(((((((.(-((((..(((....)))...)))))..-....))).))))))).......--------.------ ( -24.00) >DroYak_CAF1 127057 101 + 1 UA-AUUUCUGCAUUUUACAAGGGUGCAUAUC---UCGCCAUGGUGCUCG-GUCUAUGCCUA-GGACUUAGAGCUGC-UAGUAGUGGUUAGGCAUAAACUAUUUAACUA------ ..-.....(((((((.....)))))))..((---(.(((((((((((((-((((.......-)))))..)))).))-)....))))).))).................------ ( -22.30) >DroAna_CAF1 111515 96 + 1 UA-AUUUUUGCAUUUUACAUGGGUGCAUAUCCUAUCGCCGUGGUGCUCG-GUUUAUGCCUA-GGACUUAGGGCUAC-UAAUAGUGGUUAGGCAUAAACU--------U------ ..-......(((((....(((((.......)))))......)))))..(-(((((((((((-.........(((((-.....)))))))))))))))))--------.------ ( -26.20) >DroPer_CAF1 129188 91 + 1 UAAAUUUUUGCAUUUUACAUG-GUGCA--UC---UCGCC-UGGUGCUCG-GUCUAUGCCUAUGGGCUUAGGACUAC-UAAUAGUGAUUAGGCGUAAACU--------A------ ........((((((......)-)))))--..---.((((-(((((((.(-((((..(((....)))...)))))..-....))).))))))))......--------.------ ( -25.10) >consensus UA_AUUUUUGCAUUUUACAUG_GUGCA__UC___UCGCC_UGGUGCUCG_GUCUAUGCCUA_GGACUUAGGACUAC_UAAUAGUGGUUAGGCAUAAACU________A______ .........((((.........)))).........(((....))).....((.((((((((...(((..............)))...)))))))).))................ (-16.16 = -15.52 + -0.64)



| Location | 14,339,613 – 14,339,706 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -19.25 |
| Consensus MFE | -13.68 |
| Energy contribution | -14.16 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14339613 93 - 27905053 ------U--------AGUUUAUGCCUAAACACUACUA-GUAGCCCUAAGUCC-UAGGCAUAAAC-CGAGCACCACGGCGA---GAUAUGCACCCAUGUAAAAUGCAGAAAU-UA ------.--------.(((((((((((...(((..((-(.....))))))..-)))))))))))-...........(((.---.(((((....)))))....)))......-.. ( -19.10) >DroVir_CAF1 143373 94 - 1 CGAUUCA--------AGUUUAUGCCUAACAACUACUAGUCAAUCCUAAGUCC-UAGGCAUAGACACGCGCACCU-AAUG--------AGCAC-AUUGUAAAAUGCAAAAUU-UU .......--------.(((((((((((...(((..(((......))))))..-)))))))))))..(((....(-((((--------....)-)))).....)))......-.. ( -19.50) >DroPse_CAF1 126530 91 - 1 ------U--------AGUUUAUGCCUAAUCACUAUUA-GUAGUCCUAAGCCCAUAGGCAUAGAC-CGAGCACCA-GGCGA---GA--UGCAC-CAUGUAAAAUGCAAAAAUUUA ------.--------.(((((((((((.......(((-(.....)))).....)))))))))))-...(((...-.(((.---(.--.....-).)))....)))......... ( -19.10) >DroYak_CAF1 127057 101 - 1 ------UAGUUAAAUAGUUUAUGCCUAACCACUACUA-GCAGCUCUAAGUCC-UAGGCAUAGAC-CGAGCACCAUGGCGA---GAUAUGCACCCUUGUAAAAUGCAGAAAU-UA ------..........(((((((((((...(((..((-(.....))))))..-)))))))))))-...(((.....((((---(.........)))))....)))......-.. ( -21.40) >DroAna_CAF1 111515 96 - 1 ------A--------AGUUUAUGCCUAACCACUAUUA-GUAGCCCUAAGUCC-UAGGCAUAAAC-CGAGCACCACGGCGAUAGGAUAUGCACCCAUGUAAAAUGCAAAAAU-UA ------.--------.(((((((((((...(((..((-(.....))))))..-)))))))))))-...(((((.........))(((((....)))))....)))......-.. ( -19.60) >DroPer_CAF1 129188 91 - 1 ------U--------AGUUUACGCCUAAUCACUAUUA-GUAGUCCUAAGCCCAUAGGCAUAGAC-CGAGCACCA-GGCGA---GA--UGCAC-CAUGUAAAAUGCAAAAAUUUA ------.--------......(((((....((((...-.)))).(((.(((....))).)))..-........)-)))).---..--((((.-.........))))........ ( -16.80) >consensus ______U________AGUUUAUGCCUAACCACUACUA_GUAGCCCUAAGUCC_UAGGCAUAGAC_CGAGCACCA_GGCGA___GA__UGCAC_CAUGUAAAAUGCAAAAAU_UA ................(((((((((((...(((..............)))...)))))))))))....((......)).........((((...........))))........ (-13.68 = -14.16 + 0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:36 2006