
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,336,762 – 14,336,861 |
| Length | 99 |
| Max. P | 0.611053 |

| Location | 14,336,762 – 14,336,861 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -20.95 |
| Consensus MFE | -9.55 |
| Energy contribution | -9.83 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
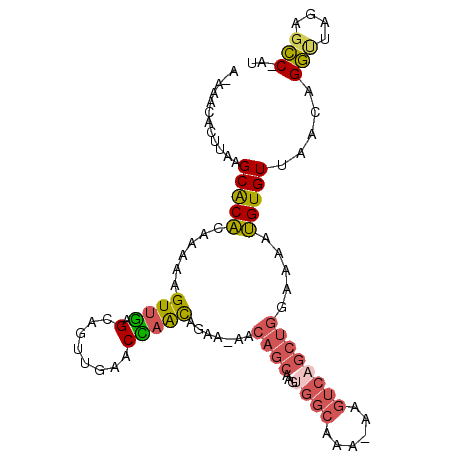
>3R_DroMel_CAF1 14336762 99 + 27905053 AAAAACAUUUAAGCACACAAAAAGUUGAGCAUUUGCACCAACAGCA-AACAGCAAGUGGCAAA-AAGUCAGCUGGAAAAUGUGUUAACAGGUUAGAGCC-AU ...........((((((......((((.((....))..))))....-..((((...((((...-..)))))))).....))))))....(((....)))-.. ( -20.60) >DroVir_CAF1 140051 92 + 1 C-CAACAAAAUGGCGCUCAAAAAGUUAAGCUGUUUAGCUUAUGCGA-AA-UGCAA---CCCCA-AAGUGU---GUAAAAAGUGUAAACAGUUUUUGCACUCG .-........(((............((((((....))))))(((..-..-.))).---..)))-..(.((---(((((((.((....)).))))))))).). ( -20.80) >DroSec_CAF1 115486 98 + 1 A-AAACACUUAAGCACACAAAAAGUUGAGCAGUUGCACCAACAGAA-AACAGCAAGUGGCAAA-AAGUCAGCUGGAAAAUGUGUUAACAGGUUAGAGCC-AU .-.........((((((......((((.((....))..))))....-..((((...((((...-..)))))))).....))))))....(((....)))-.. ( -20.60) >DroSim_CAF1 111058 99 + 1 A-AAACACUUAAGCACACAAAAAGUUGAGCAGUUGCACCAACAGAA-AACAGCAAGUGGCAAAAAAGUCAGCUGGAAAAUGUGUUAACAGGUUAGAGCC-AU .-.........((((((......((((.((....))..))))....-..((((...((((......)))))))).....))))))....(((....)))-.. ( -20.80) >DroEre_CAF1 123831 98 + 1 --AAACACUUGUGCACAUAAAAAGUUGAGCAGUUGAACUAGCAGAAAAACAGCAAGAGGCAAA-AAGUCAGCUGGAAAAUGUGUUAACAGGUUAAAGCC-AU --....((((((((((((....((((((((((.....)).)).........((.....))...-...)))))).....))))))..)))))).......-.. ( -20.80) >DroYak_CAF1 123606 98 + 1 A-AAACACUUGUGCACACAAAAAGUUGAGCAGUUGAACUAGCAGAA-CACAGCAAGUGGAAGA-AAGUCAGCUGGAAAAUGUGUUAACAGGCUAAAGCC-AU .-.(((((((((....)))).........(((((((.((..(....-(((.....)))...).-.)))))))))......)))))....(((....)))-.. ( -22.10) >consensus A_AAACACUUAAGCACACAAAAAGUUGAGCAGUUGAACCAACAGAA_AACAGCAAGUGGCAAA_AAGUCAGCUGGAAAAUGUGUUAACAGGUUAGAGCC_AU ............(((((......((((.(........))))).......((((...((((......)))))))).....))))).....(((....)))... ( -9.55 = -9.83 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:34 2006