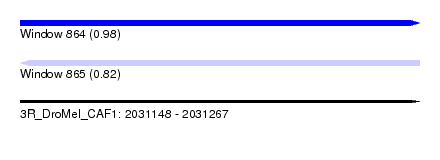
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,031,148 – 2,031,267 |
| Length | 119 |
| Max. P | 0.977884 |

| Location | 2,031,148 – 2,031,267 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -19.69 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
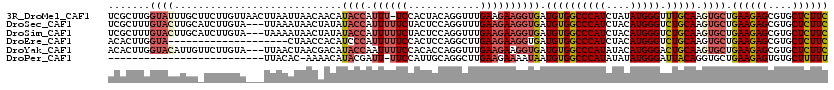
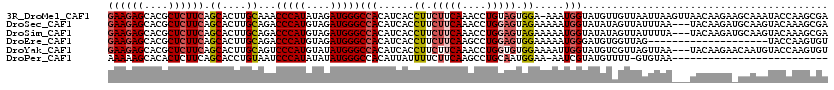
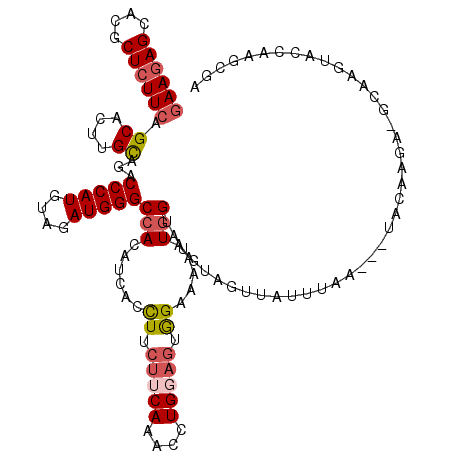
>3R_DroMel_CAF1 2031148 119 + 27905053 UCGCUUGGUAUUUGCUUCUUGUUAACUUAAUUAACAACAUACCAUUU-UCCACUACAGGUUUGAAGAAGGUGAUGUGGCCCAUCUAUAUGGGUUUGCAAGUGCUGAAGAGCGUGCUCUUC ......(((((((((....((((((.....))))))(((((((.(((-((.((.....))..))))).))).))))(((((((....))))))).)))))))))((((((....)))))) ( -35.10) >DroSec_CAF1 174163 117 + 1 UCGCUUUGUACUUGCAUCUUGUA---UUAAAUAACUAUAUACCAUUUUUCUACUCCAGGUUUGAAGAAGGUGAUGUGGCCCAUCUACAUGGGUCUGCAAGUGCUGAAGAGCGUGCUCUUC .......(((((((((.......---.............((((.((((((.((.....))..))))))))))....(((((((....)))))))))))))))).((((((....)))))) ( -37.20) >DroSim_CAF1 179101 117 + 1 UCGCUUUGUACUUGCAUCUUGUA---UAAAAUAACUAUAUACCAUUUUUCUACUCCAGGUUUGAAGAAGGUGAUGUGGCCCAUCUACAUGGGUCUGCAAGUGCUGAAGAGCGUGCUCUUC .......(((((((((.......---.............((((.((((((.((.....))..))))))))))....(((((((....)))))))))))))))).((((((....)))))) ( -37.20) >DroEre_CAF1 174785 100 + 1 ACACUUGGUA--------------------CUAACCACAUCCCAUUUUUCCACUCCAGGCUUGAAGAAGGUGAUGUGGCCCAUCUACAUGGGUCUGCAAGUGCUGAAGAGCGUGCUCUUC .((((((...--------------------....(((((((((.(((((((......))...))))).)).)))))))(((((....)))))....))))))..((((((....)))))) ( -34.60) >DroYak_CAF1 177439 117 + 1 ACACUUGGUACAUUGUUCUUGUA---UUAACUAACGACAUACCAAUUUUCCACACCAGGUUUGAAGAAGGUGAUGUGGCCCAUAUACAUGGGACUGCAAGUGCUGAAGAGCGUGCUCUUC .(((((((((..(((((...((.---...)).)))))..)))))........((((............)))).((..((((((....))))).)..))))))..((((((....)))))) ( -31.40) >DroPer_CAF1 153813 92 + 1 --------------------------UUACAC-AAAACAUACGAUU-UUCCAUUGCAGGCUUGAAGAAAAUAAUGUGGCCCAUAUAUAUGGGAUUACAGGUGCUGAAGAGUGUGCUUUUU --------------------------......-.............-.......(((.((((..((.......((((((((((....))))).)))))....))...)))).)))..... ( -20.20) >consensus UCGCUUGGUACUUGC_UCUUGUA___UUAAAUAACAACAUACCAUUUUUCCACUCCAGGUUUGAAGAAGGUGAUGUGGCCCAUCUACAUGGGUCUGCAAGUGCUGAAGAGCGUGCUCUUC .......((((............................((((.((((((............)))))))))).((((((((((....))))).))))).)))).((((((....)))))) (-19.69 = -20.67 + 0.97)
| Location | 2,031,148 – 2,031,267 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -29.39 |
| Consensus MFE | -14.83 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
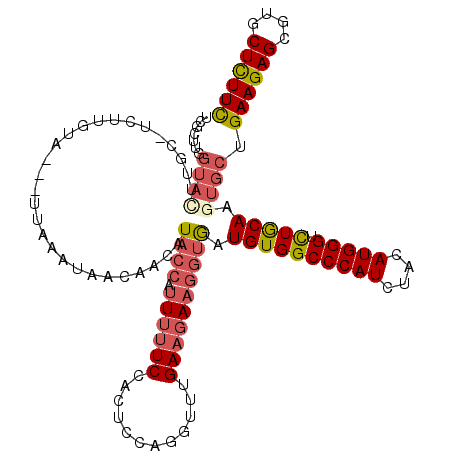
>3R_DroMel_CAF1 2031148 119 - 27905053 GAAGAGCACGCUCUUCAGCACUUGCAAACCCAUAUAGAUGGGCCACAUCACCUUCUUCAAACCUGUAGUGGA-AAAUGGUAUGUUGUUAAUUAAGUUAACAAGAAGCAAAUACCAAGCGA ((((((....)))))).....((((...(((((....)))))((((.....................)))).-...((((((.((((((((...)))))))).......)))))).)))) ( -28.30) >DroSec_CAF1 174163 117 - 1 GAAGAGCACGCUCUUCAGCACUUGCAGACCCAUGUAGAUGGGCCACAUCACCUUCUUCAAACCUGGAGUAGAAAAAUGGUAUAUAGUUAUUUAA---UACAAGAUGCAAGUACAAAGCGA ((((((....)))))).(.((((((.(.(((((....))))).)..(((..((.(((((....))))).)).......((((.(((....))))---)))..))))))))).)....... ( -29.80) >DroSim_CAF1 179101 117 - 1 GAAGAGCACGCUCUUCAGCACUUGCAGACCCAUGUAGAUGGGCCACAUCACCUUCUUCAAACCUGGAGUAGAAAAAUGGUAUAUAGUUAUUUUA---UACAAGAUGCAAGUACAAAGCGA ((((((....)))))).(.((((((.(.(((((....))))).)..(((..((.(((((....))))).)).......(((((.........))---)))..))))))))).)....... ( -31.30) >DroEre_CAF1 174785 100 - 1 GAAGAGCACGCUCUUCAGCACUUGCAGACCCAUGUAGAUGGGCCACAUCACCUUCUUCAAGCCUGGAGUGGAAAAAUGGGAUGUGGUUAG--------------------UACCAAGUGU ((((((....)))))).(((((((....(((((....)))))(((((((.((..(((((....))))).........)))))))))....--------------------...))))))) ( -36.50) >DroYak_CAF1 177439 117 - 1 GAAGAGCACGCUCUUCAGCACUUGCAGUCCCAUGUAUAUGGGCCACAUCACCUUCUUCAAACCUGGUGUGGAAAAUUGGUAUGUCGUUAGUUAA---UACAAGAACAAUGUACCAAGUGU ((((((....)))))).((((((.....(((((....)))))((((((((.............))))))))......(((((((.(((.((...---.))...))).))))))))))))) ( -35.92) >DroPer_CAF1 153813 92 - 1 AAAAAGCACACUCUUCAGCACCUGUAAUCCCAUAUAUAUGGGCCACAUUAUUUUCUUCAAGCCUGCAAUGGAA-AAUCGUAUGUUUU-GUGUAA-------------------------- .....(((((..................(((((....)))))..((((.(((((((....(....)...))))-)))...))))..)-))))..-------------------------- ( -14.50) >consensus GAAGAGCACGCUCUUCAGCACUUGCAGACCCAUGUAGAUGGGCCACAUCACCUUCUUCAAACCUGGAGUGGAAAAAUGGUAUGUAGUUAUUUAA___UACAAGA_GCAAGUACCAAGCGA ((((((....)))))).((....))...(((((....)))))(((......((.(((((....))))).)).....)))......................................... (-14.83 = -15.50 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:38 2006