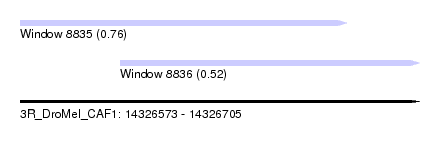
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,326,573 – 14,326,705 |
| Length | 132 |
| Max. P | 0.764383 |

| Location | 14,326,573 – 14,326,681 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.25 |
| Mean single sequence MFE | -34.78 |
| Consensus MFE | -28.09 |
| Energy contribution | -29.70 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
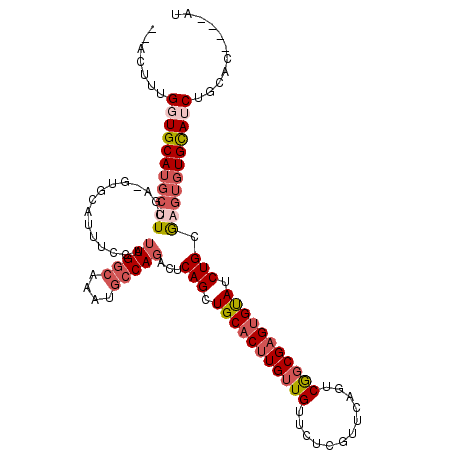
>3R_DroMel_CAF1 14326573 108 + 27905053 --ACUGUGUUGCAUGCUUUGA-GUGCAUUUCCAUUGGCAAAUGCCAGAGUCAGCUGCACUUGUUGUUCUCGUUCAGUCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC----AU --..((((((((((((((...-...........(((((....))))).(.(((.(((((((((((..((.....)).))))))))))).)))))))))))))...))))----). ( -38.00) >DroSec_CAF1 105360 108 + 1 --ACUCUGGUGCAUGCUUUGA-GUGCAUUUCCAUUGGCAAAUGCCAGAGUCAGCUGCACUUGUUGUUCUCGUUCAGUCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC----AU --(((((((((..((((..((-(.....)))....))))..)))))))))(((.(((((((((((..((.....)).))))))))))).)))...((((((....))))----)) ( -39.80) >DroSim_CAF1 100586 108 + 1 --ACUUUGGUGCAUGCUUUGA-GUGCAUUUCCAUUGGCAAAUGCCAGAGUCAGCUGCACUUGUUGUUCUCGUUCAGUCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC----AU --.....(((((((((((...-...........(((((....))))).(.(((.(((((((((((..((.....)).))))))))))).))))))))))))))).....----.. ( -37.20) >DroEre_CAF1 108090 110 + 1 UAACUUUGGUGCAUGCGUCGA-GUGCAUUCUCAUUGGCAAAUGCCAGGCUCAGCUGCACUUGUUGUUCUCGUUCAGGCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC----AU .......((((((((((((((-(......)))..((((....))))))).(((.(((((((((((((........))))))))))))).)))...))))))))).....----.. ( -41.70) >DroYak_CAF1 113060 110 + 1 UAACUUUGGUGCAUGCUUCAA-GUGCACAUCCAUUGGCAAAUGCCAGACUCAGCUGCACUUGUUGUUCUCGUUCAGUCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC----AU ........(((((.(......-(((((((..(.(((((....)))))...(((.(((((((((((..((.....)).))))))))))).))).)..)))))))))))))----.. ( -38.40) >DroAna_CAF1 96528 110 + 1 --UUCUUGUUGCAUUUUUCUACCUGUAUUCCCAUUGCC-AUUACCAUUCCCAGCUGCACUUGUUGUUCUCGUUCACUCACCGCGAGCAUCUGUA--UCUGUAUCUGCAUCGGUUU --...(((.((((...........(((.......))).-...........(((.((((.....(((((.((.........)).)))))..))))--.)))....)))).)))... ( -13.60) >consensus __ACUUUGGUGCAUGCUUCGA_GUGCAUUUCCAUUGGCAAAUGCCAGACUCAGCUGCACUUGUUGUUCUCGUUCAGUCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC____AU .......(((((((((((...............(((((....)))))...(((.(((((((((((............))))))))))).))).)))))))))))........... (-28.09 = -29.70 + 1.61)


| Location | 14,326,606 – 14,326,705 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.67 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
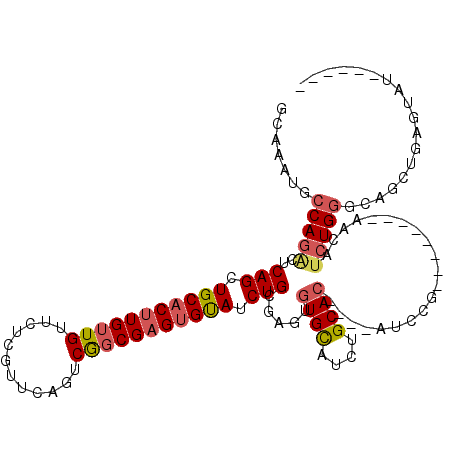
>3R_DroMel_CAF1 14326606 99 + 27905053 GCAAAUGCCAGAGUCAGCUGCACUUGUUGUUCUCGUUCAGUCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC----AUCCG--------AACAUCUGGGCAGCUGAGUAU------ .((..(((((((..(((.(((((((((((..((.....)).))))))))))).)))((..(((((....))))----)..))--------....))).))))..)).....------ ( -35.90) >DroSec_CAF1 105393 107 + 1 GCAAAUGCCAGAGUCAGCUGCACUUGUUGUUCUCGUUCAGUCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC----AUCCGAACAUCCGAACAUCUGGGCAGCUGAGUAU------ ((....))...(.((((((((.((...((((((((((.....)))))((((.....((..(((((....))))----)..)).))))..)))))...)))))))))).)..------ ( -36.80) >DroSim_CAF1 100619 99 + 1 GCAAAUGCCAGAGUCAGCUGCACUUGUUGUUCUCGUUCAGUCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC----AUCCG--------AACAUCUGGGCAGCUGAGUAU------ .((..(((((((..(((.(((((((((((..((.....)).))))))))))).)))((..(((((....))))----)..))--------....))).))))..)).....------ ( -35.90) >DroEre_CAF1 108125 105 + 1 GCAAAUGCCAGGCUCAGCUGCACUUGUUGUUCUCGUUCAGGCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC----AUCCG--------AACAUCUGGGCAUCUGAGUAUCUGAGU .((.(((((((((((((.(((((((((((((........)))))))))))))....((..(((((....))))----)..))--------.....))))))..))).)))).))... ( -39.40) >DroYak_CAF1 113095 99 + 1 GCAAAUGCCAGACUCAGCUGCACUUGUUGUUCUCGUUCAGUCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC----AUCCG--------AACAUCUGGGCAUCUAAGCAU------ ((..((((((((..(((.(((((((((((..((.....)).))))))))))).)))((..(((((....))))----)..))--------....))).)))))....))..------ ( -36.10) >DroAna_CAF1 96562 97 + 1 CC-AUUACCAUUCCCAGCUGCACUUGUUGUUCUCGUUCACUCACCGCGAGCAUCUGUA--UCUGUAUCUGCAUCGGUUUGAG--------UGCAUCUGCACAUCUGAG--------- .(-(..(((.....(((.((((..((..(..(((((.........)))))...)..))--..)))).)))....))).)).(--------(((....)))).......--------- ( -18.40) >consensus GCAAAUGCCAGACUCAGCUGCACUUGUUGUUCUCGUUCAGUCGGCGAGUGUAUCUGCGAGUGUGCAUCUGCAC____AUCCG________AACAUCUGGGCAGCUGAGUAU______ .......(((((..(((.(((((((((((............))))))))))).))).....((((....)))).....................))))).................. (-22.02 = -22.67 + 0.64)

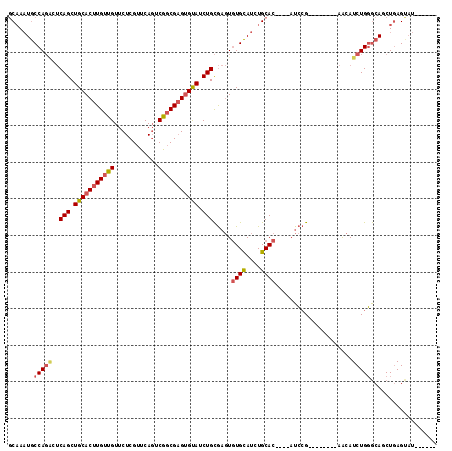
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:29 2006