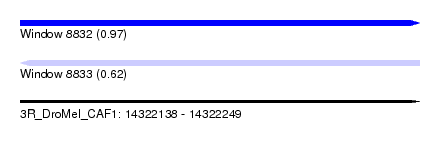
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,322,138 – 14,322,249 |
| Length | 111 |
| Max. P | 0.965518 |

| Location | 14,322,138 – 14,322,249 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -12.22 |
| Energy contribution | -13.00 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14322138 111 + 27905053 AAUAGCUAAGUCAACCCUUAAGUGGAAAC--GUAAGCCAAGUUAGCUGAGGAAACUAUCCAAAAAGUUAAUAAAUGAGCUAAUAUAGAGCAUAAACAAAGCUAAGUAAAGUAA ..(((((...(((...((((.((....))--.))))....(((((((..(((.....)))....)))))))...)))(((.......)))........))))).......... ( -19.00) >DroSec_CAF1 101229 104 + 1 AAUAGUUAAGUCAACCCUUAAGUGGGAACAACUUAGCCAAGUUAGCUGAGGGAAUCUUCCAAAAAGUGAAUAAAUGAGAU---------UCCAAACAAAGCUAAGUAAGUUAU ....(((((((...(((......)))....)))))))..(.((((((...((((((((.................)))))---------)))......)))))).)....... ( -25.13) >DroSim_CAF1 96059 98 + 1 AAUAGCUAAGUCAACCCUUAAGUGGGAAUAAGUUAGCCAAGUUAGCUGAAGGAAUCUUCCAAAAAGUUAAUUAAU---------------UCAAACAAAGCUAAUUAAGCUAU .((((((.......(((......)))....(((((((..((((((((...(((....)))....))))))))...---------------.........))))))).)))))) ( -21.64) >consensus AAUAGCUAAGUCAACCCUUAAGUGGGAACAAGUUAGCCAAGUUAGCUGAGGGAAUCUUCCAAAAAGUUAAUAAAUGAG_U__________ACAAACAAAGCUAAGUAAGCUAU ..(((((.......(((......)))..............(((((((...(((....)))....)))))))...........................))))).......... (-12.22 = -13.00 + 0.78)

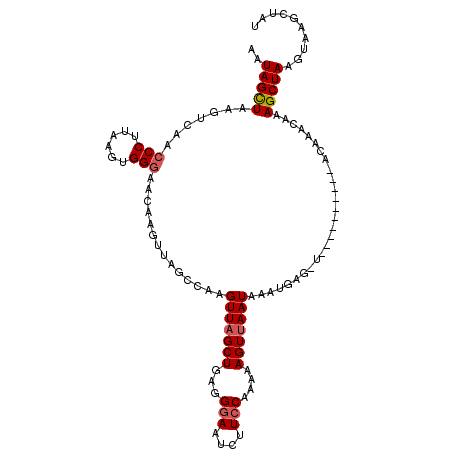

| Location | 14,322,138 – 14,322,249 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -11.57 |
| Energy contribution | -11.47 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
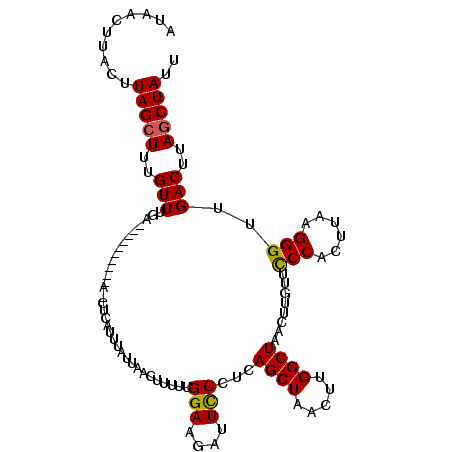
>3R_DroMel_CAF1 14322138 111 - 27905053 UUACUUUACUUAGCUUUGUUUAUGCUCUAUAUUAGCUCAUUUAUUAACUUUUUGGAUAGUUUCCUCAGCUAACUUGGCUUAC--GUUUCCACUUAAGGGUUGACUUAGCUAUU ..........(((((..(((...(((((...((((((.....((((.((....)).))))......))))))..(((.....--....)))....))))).)))..))))).. ( -19.30) >DroSec_CAF1 101229 104 - 1 AUAACUUACUUAGCUUUGUUUGGA---------AUCUCAUUUAUUCACUUUUUGGAAGAUUCCCUCAGCUAACUUGGCUAAGUUGUUCCCACUUAAGGGUUGACUUAACUAUU .........((((((..(...(((---------((((......((((.....))))))))))))..))))))..(((.(((((((..(((......))).))))))).))).. ( -22.30) >DroSim_CAF1 96059 98 - 1 AUAGCUUAAUUAGCUUUGUUUGA---------------AUUAAUUAACUUUUUGGAAGAUUCCUUCAGCUAACUUGGCUAACUUAUUCCCACUUAAGGGUUGACUUAGCUAUU .((((((((((((.(((....))---------------))))))))........((((....)))))))))...(((((((.(((..(((......))).))).))))))).. ( -21.70) >consensus AUAACUUACUUAGCUUUGUUUGA__________A_CUCAUUUAUUAACUUUUUGGAAGAUUCCCUCAGCUAACUUGGCUAACUUGUUCCCACUUAAGGGUUGACUUAGCUAUU ..........(((((..(((.................................(((....)))...((((.....))))........(((......)))..)))..))))).. (-11.57 = -11.47 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:26 2006