
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,313,801 – 14,313,891 |
| Length | 90 |
| Max. P | 0.716514 |

| Location | 14,313,801 – 14,313,891 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -20.16 |
| Consensus MFE | -15.24 |
| Energy contribution | -15.60 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
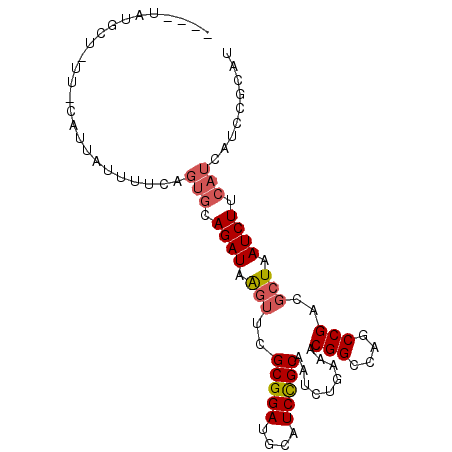
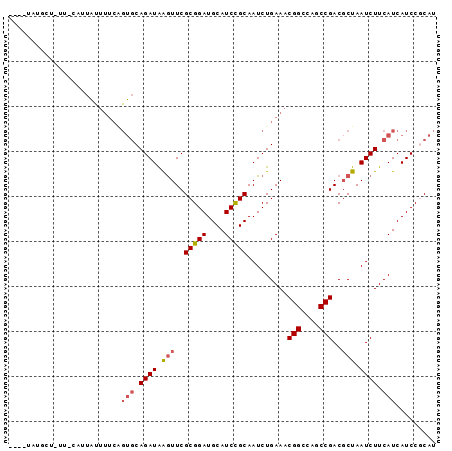
>3R_DroMel_CAF1 14313801 90 + 27905053 ----UAUGCUGUU-CAUUAUUUUCAGUGCAGAUAGAUUCGCGGAUGCAUCCGCAAUCUGAAACGGCCAGCCGACGCUAAUCUUCAUCAUCCGCAU ----...((((..-.........))))((.((((((((.(((((....))))))))))....(((....)))...............))).)).. ( -21.60) >DroSec_CAF1 92925 90 + 1 ----UAUGCUAUU-CAUUAUUUUCAGUGCAGAUGGGUUCGCGGAUGCAUCCGCAAUCUGAAACGGCCAGCCGACGCUAAUCUUCAUCAUCCGCAU ----.((((....-......(((((((((.((((.(((....))).)))).)))..))))))(((....)))...................)))) ( -23.10) >DroSim_CAF1 86718 90 + 1 ----UAUGCUAUU-CAUUAUUUUCAGUGCAGAUAGGUUCGCGGAUGCAUCCGCAAUCUGAAACGGCCAGCCGACGCUAAUCUUCAUCAUCCGCAU ----.........-...........((((.((((((((.(((((....))))))))))....(((....)))...............))).)))) ( -20.60) >DroEre_CAF1 95805 86 + 1 CAC--------CCAAAUUGCUUUCGGUGCAGAUAAGUUCGCGGAUGCAUCUGCAAUCUGAAACGGCCAGCCGACUCUAAUCUUCAUCAUCC-CAU ...--------.........(((((((((((((..(((....)))..)))))))..))))))(((....)))...................-... ( -18.90) >DroYak_CAF1 93496 94 + 1 CACUUAUCCACUUAAAUUACUUUCUGUGCAGAUAAGUUCGCGGAUGCAUCCGCAAUCUGAAACGGCCAGCCGACGCUAAUCUUCAUCAUCC-CAU .((((((((((..(((....)))..)))..)))))))..(((((....))))).........(((....)))...................-... ( -19.30) >DroAna_CAF1 84795 68 + 1 ---------------------------CCAGAUAAGUUCGCGGAUGCAUCUGCAAUCUGAAUCGGCCUGCCGACGCUAAUCUUCAUCAUCCGCAU ---------------------------............(((((((............(..((((....))))..)..........))))))).. ( -17.45) >consensus ____UAUGCU_UU_CAUUAUUUUCAGUGCAGAUAAGUUCGCGGAUGCAUCCGCAAUCUGAAACGGCCAGCCGACGCUAAUCUUCAUCAUCCGCAU .........................(((.((((.(((..(((((....))))).........(((....)))..))).)))).)))......... (-15.24 = -15.60 + 0.36)

| Location | 14,313,801 – 14,313,891 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -18.52 |
| Energy contribution | -19.88 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
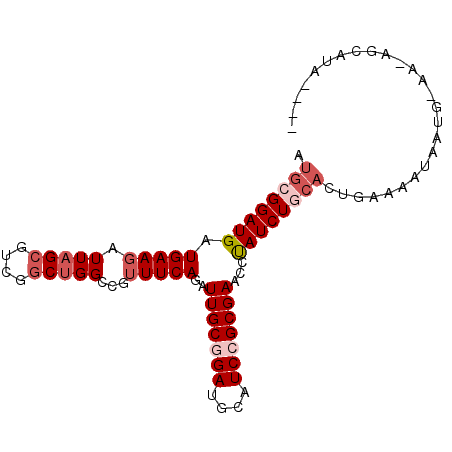
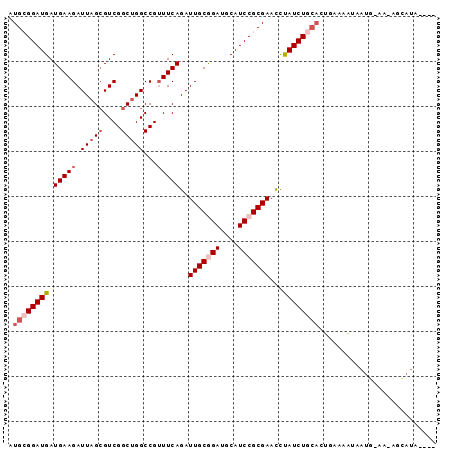
>3R_DroMel_CAF1 14313801 90 - 27905053 AUGCGGAUGAUGAAGAUUAGCGUCGGCUGGCCGUUUCAGAUUGCGGAUGCAUCCGCGAAUCUAUCUGCACUGAAAAUAAUG-AACAGCAUA---- .((((((((.(((((.(((((....)))))...)))))(((((((((....))))).))))))))))))............-.........---- ( -26.50) >DroSec_CAF1 92925 90 - 1 AUGCGGAUGAUGAAGAUUAGCGUCGGCUGGCCGUUUCAGAUUGCGGAUGCAUCCGCGAACCCAUCUGCACUGAAAAUAAUG-AAUAGCAUA---- .((((((((.(((((.(((((....)))))...)))))..(((((((....)))))))...))))))))............-.........---- ( -27.80) >DroSim_CAF1 86718 90 - 1 AUGCGGAUGAUGAAGAUUAGCGUCGGCUGGCCGUUUCAGAUUGCGGAUGCAUCCGCGAACCUAUCUGCACUGAAAAUAAUG-AAUAGCAUA---- .((((((((.(((((.(((((....)))))...)))))..(((((((....)))))))...))))))))............-.........---- ( -25.70) >DroEre_CAF1 95805 86 - 1 AUG-GGAUGAUGAAGAUUAGAGUCGGCUGGCCGUUUCAGAUUGCAGAUGCAUCCGCGAACUUAUCUGCACCGAAAGCAAUUUGG--------GUG ...-(((((.((..((((.(((.(((....))).))).)))).))....))))).............((((.(((....))).)--------))) ( -23.00) >DroYak_CAF1 93496 94 - 1 AUG-GGAUGAUGAAGAUUAGCGUCGGCUGGCCGUUUCAGAUUGCGGAUGCAUCCGCGAACUUAUCUGCACAGAAAGUAAUUUAAGUGGAUAAGUG .((-..((((((........))))((....))))..))..(((((((....)))))))(((((((..(....(((....)))..)..))))))). ( -28.80) >DroAna_CAF1 84795 68 - 1 AUGCGGAUGAUGAAGAUUAGCGUCGGCAGGCCGAUUCAGAUUGCAGAUGCAUCCGCGAACUUAUCUGG--------------------------- .((((((((.((..((((.(.(((((....))))).).)))).))....))))))))...........--------------------------- ( -22.30) >consensus AUGCGGAUGAUGAAGAUUAGCGUCGGCUGGCCGUUUCAGAUUGCGGAUGCAUCCGCGAACCUAUCUGCACUGAAAAUAAUG_AA_AGCAUA____ .((((((((.(((((.(((((....)))))...)))))..(((((((....)))))))...)))))))).......................... (-18.52 = -19.88 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:23 2006