
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,279,062 – 14,279,153 |
| Length | 91 |
| Max. P | 0.933087 |

| Location | 14,279,062 – 14,279,153 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -16.58 |
| Consensus MFE | -13.01 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
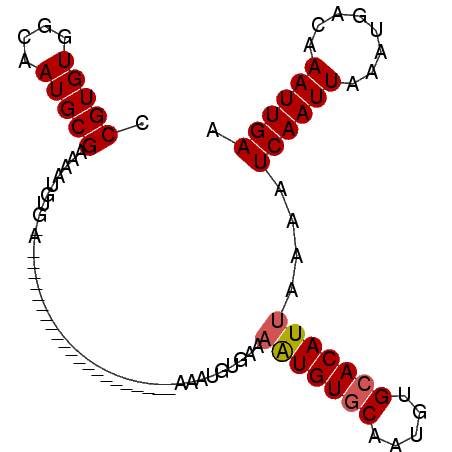
>3R_DroMel_CAF1 14279062 91 + 27905053 CCGUGUGGCAAUGCGAAAAUGUG-AAUGUGAAAAU-------GUGGAAAUGUGAAAAUGUGCAAUGUGCACAUUAAAAUCAAUUAAAUGACAAAUUGAA (((..(..((...((......))-....))...).-------.))).........(((((((.....)))))))....((((((........)))))). ( -15.70) >DroSec_CAF1 58433 69 + 1 CCGUGUGGCAAUGCGAAAA------------------------------UGUGGAAAUGUGCAAUGUGCACAUUAAAAUCAAUUAAAUGACAAAUUGAA .(((((....)))))....------------------------------......(((((((.....)))))))....((((((........)))))). ( -14.60) >DroSim_CAF1 50121 77 + 1 CCGUGUGGCAAUGCGAAAAUGUG-A---------------------AAAUGUGGAAAUGUGCAAUGUGCACAUUAAAAUCAAUUAAAUGACAAAUUGAA .(((((....)))))...((((.-.---------------------..))))...(((((((.....)))))))....((((((........)))))). ( -14.90) >DroEre_CAF1 60567 98 + 1 CCGUGUGGCAAUGCGAAAAUGUGCAAUGUGGAAAUGUGCAAUGUGGAAAUGUGC-AAUGUGCAAUGUGCACAUUAAAAUCAAUUAAAUGACAAAUUGAA .(((((....)))))..((((((((...((.(....((((.(((....))))))-)...).))...))))))))....((((((........)))))). ( -24.10) >DroYak_CAF1 55389 76 + 1 CCGUGUGGCAAUGCGAAAAU----------------------GUGGAAGUAUGC-AAUGUGCAAUGUGCACAUUAAAAUCAAUUAAAUGACAAAUUGAA ..((((((((.((((....(----------------------(((......)))-)...)))).))).))))).....((((((........)))))). ( -14.90) >DroAna_CAF1 50199 77 + 1 CCGUGUGGAAAUGCGAAAAUGUG-A---------------------AAAUGCGACUGUGUUCGGUGUGCACAUUAAAAUCAAUUAAAUGACAAAUUGAA .(((((....)))))..((((((-.---------------------....((.((((....)))))).))))))....((((((........)))))). ( -15.30) >consensus CCGUGUGGCAAUGCGAAAAUGUG_A_____________________AAAUGUGAAAAUGUGCAAUGUGCACAUUAAAAUCAAUUAAAUGACAAAUUGAA .(((((....)))))........................................(((((((.....)))))))....((((((........)))))). (-13.01 = -13.45 + 0.44)


| Location | 14,279,062 – 14,279,153 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -11.20 |
| Consensus MFE | -8.82 |
| Energy contribution | -9.32 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14279062 91 - 27905053 UUCAAUUUGUCAUUUAAUUGAUUUUAAUGUGCACAUUGCACAUUUUCACAUUUCCAC-------AUUUUCACAUU-CACAUUUUCGCAUUGCCACACGG .((((((........))))))....(((((((.....))))))).............-------...........-....................... ( -10.70) >DroSec_CAF1 58433 69 - 1 UUCAAUUUGUCAUUUAAUUGAUUUUAAUGUGCACAUUGCACAUUUCCACA------------------------------UUUUCGCAUUGCCACACGG .((((((........))))))....(((((((.....)))))))......------------------------------................... ( -10.70) >DroSim_CAF1 50121 77 - 1 UUCAAUUUGUCAUUUAAUUGAUUUUAAUGUGCACAUUGCACAUUUCCACAUUU---------------------U-CACAUUUUCGCAUUGCCACACGG .((((((........))))))....(((((((.....))))))).........---------------------.-....................... ( -10.70) >DroEre_CAF1 60567 98 - 1 UUCAAUUUGUCAUUUAAUUGAUUUUAAUGUGCACAUUGCACAUU-GCACAUUUCCACAUUGCACAUUUCCACAUUGCACAUUUUCGCAUUGCCACACGG .((((((........))))))...((((((((.....)))))))-).........................((.(((........))).))........ ( -15.30) >DroYak_CAF1 55389 76 - 1 UUCAAUUUGUCAUUUAAUUGAUUUUAAUGUGCACAUUGCACAUU-GCAUACUUCCAC----------------------AUUUUCGCAUUGCCACACGG .((((((........))))))...((((((((.....)))))))-)...........----------------------.................... ( -12.10) >DroAna_CAF1 50199 77 - 1 UUCAAUUUGUCAUUUAAUUGAUUUUAAUGUGCACACCGAACACAGUCGCAUUU---------------------U-CACAUUUUCGCAUUUCCACACGG .((((((........))))))....((((((.....(((......))).....---------------------.-))))))................. ( -7.70) >consensus UUCAAUUUGUCAUUUAAUUGAUUUUAAUGUGCACAUUGCACAUUUCCACAUUU_____________________U_CACAUUUUCGCAUUGCCACACGG .((((((........))))))....(((((((.....)))))))....................................................... ( -8.82 = -9.32 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:14 2006