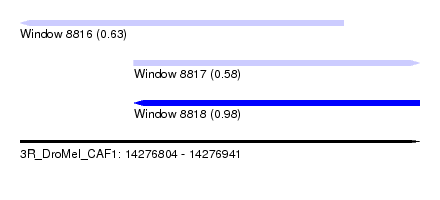
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,276,804 – 14,276,941 |
| Length | 137 |
| Max. P | 0.981537 |

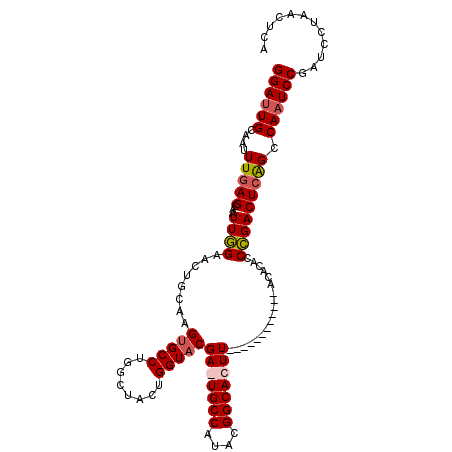
| Location | 14,276,804 – 14,276,915 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 85.20 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -22.65 |
| Energy contribution | -23.53 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14276804 111 - 27905053 GGAUUGCAAUUUGAGCAAUUGGAACUGCAAGUGCCUGGCUACUGGUACGA-UGCCAUACGGCACUUACAUACUAACUUACACACCCGACUCAGCCAAUCCGAUCCUAACUCA ((((((....(((((...((((........(((((........)))))..-((((....)))).....................))))))))).))))))............ ( -26.10) >DroSec_CAF1 56104 100 - 1 GGAUUGCAAUUUGAGCAAUUGGAACUGCAAGUGCCUGGCUACUGGUACGAGUGCCAUACGGCACUU------------ACACACCCGACUCGGCCAAUCCGAUCCUAACUCA ((((((....(((.((..((((...((.(((((((((((.(((......)))))))...)))))))------------.))...))))....)))))..))))))....... ( -32.30) >DroSim_CAF1 47809 100 - 1 GGAUUGCAAUUUAAGCAAUUGGAACUGCAAGUGCCUUGCUACUGGUACGAGUGCCAUACGGCACUU------------ACACACCCGACUCAGCCAAUCCGAUCCUAACUCA (((((((.......))))(((((...((..(((((........)))))(((((((....)))))))------------..............))...))))))))....... ( -30.00) >DroYak_CAF1 52964 89 - 1 GGACUGCAAUUUGAGCAAUUUGAACUGGAAGUGCCUGGCUACUGGUUCGA-U----------ACUU------------ACACACCAGACUCAGCCAAUCCGAUCCUAACUCA (((..(((.(((.((.........)).))).))).(((((.(((((....-.----------....------------....)))))....))))).)))............ ( -17.42) >consensus GGAUUGCAAUUUGAGCAAUUGGAACUGCAAGUGCCUGGCUACUGGUACGA_UGCCAUACGGCACUU____________ACACACCCGACUCAGCCAAUCCGAUCCUAACUCA ((((((....(((((...((((........(((((........)))))(((((((....)))))))..................))))))))).))))))............ (-22.65 = -23.53 + 0.88)

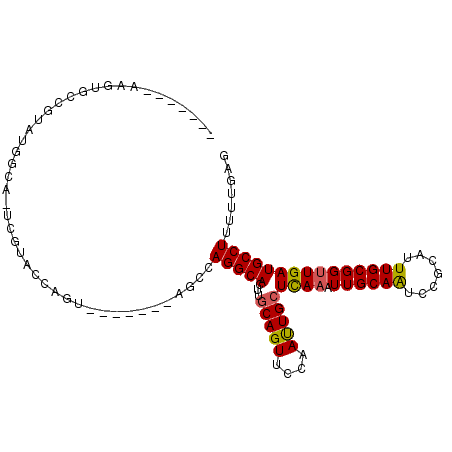
| Location | 14,276,843 – 14,276,941 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.08 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14276843 98 + 27905053 AGUAUGUAAGUGCCGUAUGGCA-UCGUACCAGU-------AGCCAGGCACUUGCAGUUCCAAUUGCUCAAAUUGCAAUCCACAUUUGCGGUUGAUGCCUUUUUGAG ....((((((((((...((((.-.(......).-------.))))))))))))))..........((((((..((((.......))))(((....)))..)))))) ( -30.70) >DroSec_CAF1 56138 92 + 1 -------AAGUGCCGUAUGGCACUCGUACCAGU-------AGCCAGGCACUUGCAGUUCCAAUUGCUCAAAUUGCAAUCCGCAUUUGCGGCUGAUGCCUUUUUGAG -------(((((((...(((((((......)))-------.)))))))))))(((((....)))))(((((..(((..((((....))))....)))...))))). ( -31.50) >DroSim_CAF1 47843 92 + 1 -------AAGUGCCGUAUGGCACUCGUACCAGU-------AGCAAGGCACUUGCAGUUCCAAUUGCUUAAAUUGCAAUCCGCAUUUGCGGUUGAUGCCUUUUUGAG -------.((((((....))))))........(-------((.((((((...(((((....)))))........(((.((((....))))))).)))))).))).. ( -27.80) >DroEre_CAF1 58264 87 + 1 -------G-GC----------A-UCGAACCAGUAGCCAGUAGCCAGGCACUUGCAGUUCAAACUGCUCAAAUUGCAAUCAGCAUUUGCGGUUGAUGCCUUUUUGAG -------(-((----------(-((.(((((((.(((........)))))).(((((....))))).......((((.......))))))))))))))........ ( -29.30) >DroYak_CAF1 52998 81 + 1 -------AAGU----------A-UCGAACCAGU-------AGCCAGGCACUUCCAGUUCAAAUUGCUCAAAUUGCAGUCCGCAUUUGCGGUUGAUGCCUUUUUGAG -------....----------.-..((((.(((-------.((...)))))....))))......((((((..(((..((((....))))....)))...)))))) ( -19.00) >consensus _______AAGUGCCGUAUGGCA_UCGUACCAGU_______AGCCAGGCACUUGCAGUUCCAAUUGCUCAAAUUGCAAUCCGCAUUUGCGGUUGAUGCCUUUUUGAG ............................................(((((...(((((....)))))((((.((((((.......)))))))))))))))....... (-15.16 = -15.08 + -0.08)

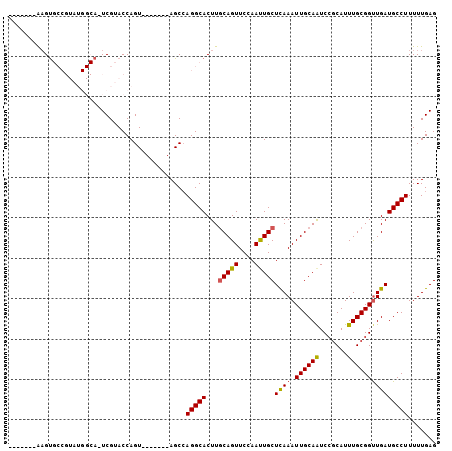
| Location | 14,276,843 – 14,276,941 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14276843 98 - 27905053 CUCAAAAAGGCAUCAACCGCAAAUGUGGAUUGCAAUUUGAGCAAUUGGAACUGCAAGUGCCUGGCU-------ACUGGUACGA-UGCCAUACGGCACUUACAUACU ........((((((..((((....)))).(((((.((..(....)..))..)))))(((((.....-------...)))))))-)))).................. ( -30.10) >DroSec_CAF1 56138 92 - 1 CUCAAAAAGGCAUCAGCCGCAAAUGCGGAUUGCAAUUUGAGCAAUUGGAACUGCAAGUGCCUGGCU-------ACUGGUACGAGUGCCAUACGGCACUU------- .(((((...(((....((((....))))..)))..)))))(((........)))..(((((.....-------...)))))(((((((....)))))))------- ( -33.40) >DroSim_CAF1 47843 92 - 1 CUCAAAAAGGCAUCAACCGCAAAUGCGGAUUGCAAUUUAAGCAAUUGGAACUGCAAGUGCCUUGCU-------ACUGGUACGAGUGCCAUACGGCACUU------- ......(((((((...((((....)))).(((((.(((((....)))))..))))))))))))...-------........(((((((....)))))))------- ( -32.10) >DroEre_CAF1 58264 87 - 1 CUCAAAAAGGCAUCAACCGCAAAUGCUGAUUGCAAUUUGAGCAGUUUGAACUGCAAGUGCCUGGCUACUGGCUACUGGUUCGA-U----------GC-C------- ........((((((((((.(((((((.....)).))))).(((((....))))).((((((........))).))))))).))-)----------))-)------- ( -30.90) >DroYak_CAF1 52998 81 - 1 CUCAAAAAGGCAUCAACCGCAAAUGCGGACUGCAAUUUGAGCAAUUUGAACUGGAAGUGCCUGGCU-------ACUGGUUCGA-U----------ACUU------- ((((((...(((....((((....))))..)))..))))))....((((((..(.(((.....)))-------.)..))))))-.----------....------- ( -24.90) >consensus CUCAAAAAGGCAUCAACCGCAAAUGCGGAUUGCAAUUUGAGCAAUUGGAACUGCAAGUGCCUGGCU_______ACUGGUACGA_UGCCAUACGGCACUU_______ ((((((...(((....((((....))))..)))..))))))...............(((((...............)))))...((((....)))).......... (-16.92 = -17.70 + 0.78)

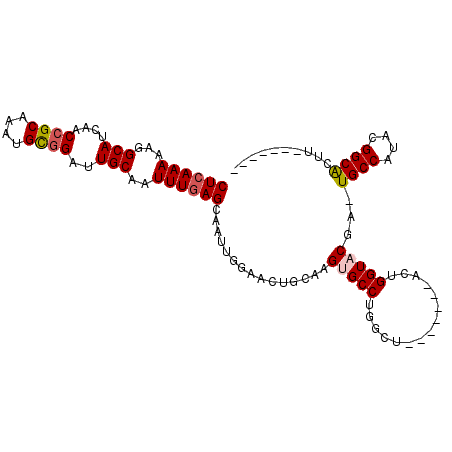
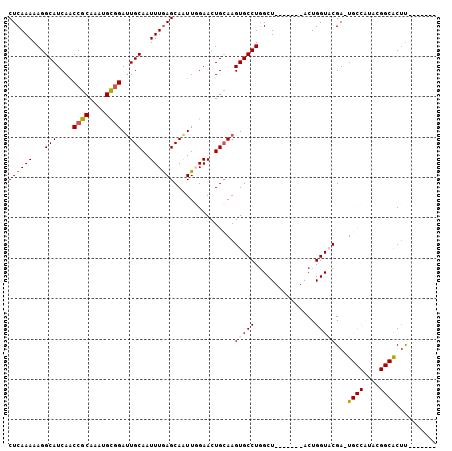
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:12 2006