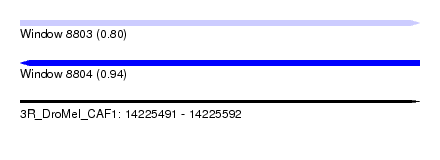
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,225,491 – 14,225,592 |
| Length | 101 |
| Max. P | 0.939095 |

| Location | 14,225,491 – 14,225,592 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 68.08 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -15.45 |
| Energy contribution | -15.57 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14225491 101 + 27905053 UAUUAAAAGCUG--------UUACC-AAAUUAUGUCCUACGCCGGCUAUAGCCAGCUGCCCAGCGGCA---GCGACCAGGAGCGCAGGCAGGCGCAGCUCGCGGCCAGCACCU ........((((--------.....-......((.(((.(((((((....))).((((((....))))---))......).))).)))))(.(((.....))).))))).... ( -35.90) >DroVir_CAF1 104473 112 + 1 UAAUCCUAUUUUACAGCCCAAAGCC-GCAGUAUGUCCUAUAAUGGGUACACCCAACUGCCUAGCGGUAGCAUCGACUACGAGCGCCGCCAGCAACAGGUGGUGGCCAACACCU .........................-((.(((.(((......((((....)))).(((((....)))))....))))))..))(((((((.(....).)))))))........ ( -32.50) >DroPse_CAF1 106136 90 + 1 UGUCAA--------------------AAAACAUGUCAUACGCCGGCUAUAGCCAGCUGCCCAGCGGCA---GCGACCACGAGCAGCGCCAGCAGCAGCUGGCUGCGAACACUU ..((..--------------------.......(((.......(((....))).((((((....))))---))))).....(((..((((((....)))))))))))...... ( -33.70) >DroYak_CAF1 97931 102 + 1 UGUUGGAAAUUC--------UUAACAAAAUUAUGUCGUAUGCCGGCUAUAGCCAGCUGCCCAGUGGCA---GCGACCAGGAGCGCAGACAGGCGCAGCUCGCGGCCAGCACCU ((((((......--------...(((......)))(((..((.(((....))).((((((....))))---))........((((......)))).))..))).))))))... ( -34.20) >DroAna_CAF1 90576 104 + 1 UACUCUAAAUUU--------AUCUG-AAAGGAUGUCCAACGCCGGCUACAGCCAGCUGCCCAUUGGCAGUGGCGACCACGAGCGACGACAGGCUCAGUUGGUGGCCAACACCU ...........(--------((((.-...))))).....(((((((....))).((((((....)))))))))).....((((........))))....((((.....)))). ( -37.30) >DroPer_CAF1 112018 90 + 1 UGUCAA--------------------AAAACAUGUCAUACGCCGGCUACAGCCAGCUGCCCAGCGGCA---GCGACCACGAGCAGCGCCAGCAGCAGCUGGCUGCGAACACUU ..((..--------------------.......(((.......(((....))).((((((....))))---))))).....(((..((((((....)))))))))))...... ( -33.70) >consensus UAUUAAAA_UU__________U__C_AAAAUAUGUCCUACGCCGGCUACAGCCAGCUGCCCAGCGGCA___GCGACCACGAGCGCCGCCAGCAGCAGCUGGCGGCCAACACCU ................................(((....((((((((.......(((((...)))))....((........))............))))))))....)))... (-15.45 = -15.57 + 0.12)



| Location | 14,225,491 – 14,225,592 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 68.08 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -20.08 |
| Energy contribution | -19.53 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14225491 101 - 27905053 AGGUGCUGGCCGCGAGCUGCGCCUGCCUGCGCUCCUGGUCGC---UGCCGCUGGGCAGCUGGCUAUAGCCGGCGUAGGACAUAAUUU-GGUAA--------CAGCUUUUAAUA ....((((((((((.....))).((((((((((.......((---((((....)))))).(((....))))))))))).))......-)))..--------))))........ ( -41.30) >DroVir_CAF1 104473 112 - 1 AGGUGUUGGCCACCACCUGUUGCUGGCGGCGCUCGUAGUCGAUGCUACCGCUAGGCAGUUGGGUGUACCCAUUAUAGGACAUACUGC-GGCUUUGGGCUGUAAAAUAGGAUUA .((((.....)))).((((((.(((((((.(((((....))).))..)))))))(((((((((....))))...........)))))-(((.....)))....)))))).... ( -39.10) >DroPse_CAF1 106136 90 - 1 AAGUGUUCGCAGCCAGCUGCUGCUGGCGCUGCUCGUGGUCGC---UGCCGCUGGGCAGCUGGCUAUAGCCGGCGUAUGACAUGUUUU--------------------UUGACA ..((((((((.((((((....))))))((((((((((((...---.))))).)))))))((((....)))))))...)))))(((..--------------------..))). ( -42.70) >DroYak_CAF1 97931 102 - 1 AGGUGCUGGCCGCGAGCUGCGCCUGUCUGCGCUCCUGGUCGC---UGCCACUGGGCAGCUGGCUAUAGCCGGCAUACGACAUAAUUUUGUUAA--------GAAUUUCCAACA ..((((((((...((((.(((......))))))).(((((((---((((....)))))).)))))..))))))))..((((......))))..--------............ ( -39.10) >DroAna_CAF1 90576 104 - 1 AGGUGUUGGCCACCAACUGAGCCUGUCGUCGCUCGUGGUCGCCACUGCCAAUGGGCAGCUGGCUGUAGCCGGCGUUGGACAUCCUUU-CAGAU--------AAAUUUAGAGUA .(((((((((.((((...((((........)))).)))).)))..((((....))))((((((....))))))....))))))....-.....--------............ ( -37.50) >DroPer_CAF1 112018 90 - 1 AAGUGUUCGCAGCCAGCUGCUGCUGGCGCUGCUCGUGGUCGC---UGCCGCUGGGCAGCUGGCUGUAGCCGGCGUAUGACAUGUUUU--------------------UUGACA ..((((((((.((((((....))))))((((((((((((...---.))))).)))))))((((....)))))))...)))))(((..--------------------..))). ( -42.70) >consensus AGGUGUUGGCCGCCAGCUGCGCCUGGCGGCGCUCGUGGUCGC___UGCCGCUGGGCAGCUGGCUAUAGCCGGCGUAGGACAUAAUUU_G__A__________AA_UUUUAACA .((((.(((.(((.....))).)))....))))....(((.....((((....))))((((((....))))))....)))................................. (-20.08 = -19.53 + -0.55)

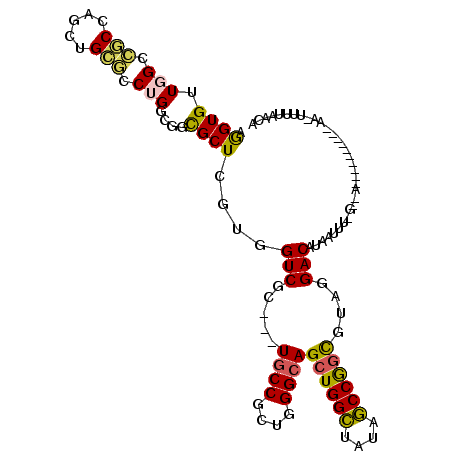
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:58 2006