
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,195,690 – 14,195,810 |
| Length | 120 |
| Max. P | 0.533055 |

| Location | 14,195,690 – 14,195,810 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.08 |
| Mean single sequence MFE | -41.01 |
| Consensus MFE | -19.69 |
| Energy contribution | -19.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14195690 120 + 27905053 AAAAUCCGCUGUAAAGGAAUAUGUGGCCGAGGCAGGAUAGUGACCGGGACUCAGCAGCGGUGGGUAUAGCACAUUUACCGCCGCUGCUGCUGCUGCCGCCGAGGUGGAGUUACUGGGUGG ...((((.((((...((.........))...))))...(((((((((.(..((((((((((((..............)))))))))))).).)))((((....)))).)))))))))).. ( -46.94) >DroVir_CAF1 75892 102 + 1 AAAGUCAGCUGUAAAGGAAUAGGUGGCCGAGGCUGGAUAGUGCGUAGGACUCAAUAGCGGCGGAUAGAGCACAUUGACAGCCGCCG------------------AGGAGUUGCUCGGUGG .....((.((((......)))).))((((((((........))....(((((.....((((((...((.....)).....))))))------------------..))))).)))))).. ( -32.70) >DroWil_CAF1 64973 111 + 1 AAAAUCCGCCGUAAAUGAAUAGGUAGCCGAGGCGGGAUAAUGGGUGGGACUUAGUAGCGGCGGAUAGAGCACAUUUACCGCUGCU------GCAGCCGCC---GUGGAAUUAUUUGGCGG .....(((((......((((((....(((.(((((.....((((.....))))((((((((((.((((.....))))))))))))------))..)))))---.)))..))))))))))) ( -45.30) >DroMoj_CAF1 79690 102 + 1 AAAGUCGGCCGUGAAGGAGUAGGUGGCCGAGGCCGGAUAGUGCGUGGGACUCAGUAGCGGCGGAUAGAGCACAUUGACCGCCGCCG------------------AGGAGCUGUUCGGCGG ....(((((((............))))))).((((((((((........(((....(((((((.(((......))).))))))).)------------------))..)))))))))).. ( -41.30) >DroAna_CAF1 62899 117 + 1 AAAGUCCGCCGUAAACGAGUAGGUAGCUGAGGCAGGAUAGUGGCCCGGACUUAGAAGUGGCGGAUAUAACACAUUAACCGCCGCCGC---AGCUGCCGCAGAAGUUGAGUUGCCGGGCGG .....(((((...........((((((((.(((.((........))............(((((.((........)).)))))))).)---)))))))((((........))))..))))) ( -45.00) >DroPer_CAF1 77765 99 + 1 GAAGUCCGCCGUGAAAGAGUAGGUGGCCGAGGCAGGGUAGUGGGUGGGACUCAGCAGCGGCGGGUAUAGCACAUUCACCGCCG---------------------AGGAACUGCUGGGUGG ....((((((.(....)....(.((.((......)).)).).))))))(((((((((((((((..............))))))---------------------.....))))))))).. ( -34.84) >consensus AAAGUCCGCCGUAAAGGAAUAGGUGGCCGAGGCAGGAUAGUGGGUGGGACUCAGUAGCGGCGGAUAGAGCACAUUGACCGCCGCCG__________________AGGAGUUGCUGGGCGG .....(((((...............((....)).......................(((((((..............)))))))...............................))))) (-19.69 = -19.47 + -0.22)

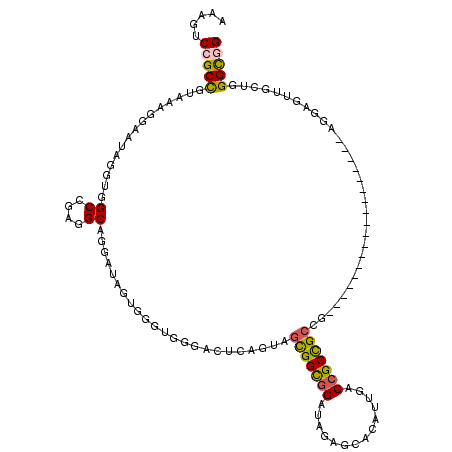
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:48 2006