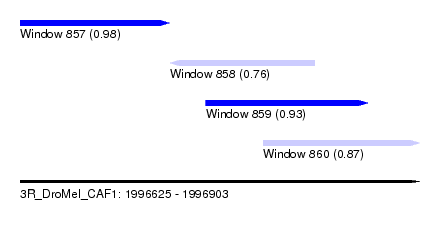
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,996,625 – 1,996,903 |
| Length | 278 |
| Max. P | 0.983231 |

| Location | 1,996,625 – 1,996,729 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.93 |
| Mean single sequence MFE | -32.74 |
| Consensus MFE | -15.68 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1996625 104 + 27905053 ACAUUCUACAUUAU-UGCGUGCCGCCUUCUUGCGGAAGCUGAUAGAAACACUUGUCCAGUGGCACCUCUU---GGAUUUAUGU----AUCUCUACAUUUAGACGGCGCUAUU-------- ..............-...((((((.((((..((....)).)).(((.(((...((((((.(....).).)---))))...)))----.)))........)).))))))....-------- ( -25.20) >DroSec_CAF1 145742 112 + 1 ACAUUCUACAUUUU-UGCCUGCUGCCAUCUCGCGGAAGCGAGGAGAAGAACUUGUCCAGUGGCACCUCUA---GUAUUUAUGU----GACUUUUCAUCUAGAUGGCGGUAUUUGAGCAAG .............(-((((((((((((((((((....))))..(((.(((...(((((...((.......---)).....)).----)))..))).))).))))))))))...).)))). ( -33.80) >DroSim_CAF1 150955 103 + 1 ACAUUCUACAUUCU----------ACAUCUUGCGGAAGCGAGGAGAAUAACUUGUCCAGUAGCACCACUA---GUAUUUAUGU----GACUUUUCAUCUAGAUGGCGCUAUUUGAGCAAG .........(((((----------...((((((....)))))))))))..((((..(((((((.((((((---(.......((----((....)))))))).))).)))).)))..)))) ( -31.91) >DroEre_CAF1 149012 91 + 1 ACAUUCUACAUUGUGUGCCUAUCGCCAUCUUGCGGAAGCAGACAGAAGCACGUGGCAAGUGUCGCCUCU---------------------UGUACAGGUAGAUGGCGGUACU-------- ((((........))))...((((((((((((((....)))((((...((.....))...))))((((..---------------------.....)))))))))))))))..-------- ( -30.30) >DroYak_CAF1 152882 112 + 1 GCAUUCUACAUCAUGUGCCUAUCGCCAUCUUGCGGAAGCAGACAGAAACACUUGUCGAGUGGCUCUGCUACUUAGGUGUUUGUAUCGGAAGGUACAUGUAGAUGGCGCUACU-------- ((((..........))))....(((((((((((....))).(((.((((((((...(((((((...)))))))))))))))(((((....))))).))))))))))).....-------- ( -42.50) >consensus ACAUUCUACAUUAU_UGCCUACCGCCAUCUUGCGGAAGCAGACAGAAACACUUGUCCAGUGGCACCUCUA___GGAUUUAUGU____GACUGUACAUCUAGAUGGCGCUAUU________ ......................(((((((((((....))))............(((....))).....................................)))))))............. (-15.68 = -15.92 + 0.24)

| Location | 1,996,729 – 1,996,830 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.03 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.10 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1996729 101 - 27905053 UCGACUGUAUGUGAAGCGAAUUCGAUUGGUAAAAACAAACCAAAUUCGCCUGAAUUUGAAGCGAAUUCGAAUCUUUUGAGUUUACAUUAUAAACUGUACGU--------------- ......((((..((..((((((((.(((.......)))..(((((((....)))))))...))))))))..)).....((((((.....))))))))))..--------------- ( -22.30) >DroSec_CAF1 145854 116 - 1 UCGACUAUAUGUGAAGCGAGUUCGAUUGGUAAAAUUAAACAGAAUUUGCCUGUAUGUGAAGCGAAUUCGAAUGGUUUGAGUUUACAUCAUCGCAUGUAGUUUUACUCGCAUGUGUG ..((((((((((((.((((((((..(((((...)))))...)))))))).((.(((((((.((((((.....))))))..))))))))))))))))))))).....(((....))) ( -30.80) >DroSim_CAF1 151058 116 - 1 UCGACUAUAUGUGAAGCGAAUUCGAUUGGUAAAAUUAAACAGAAUUUGCCUGUAUGUGAAGCGAAUUCGAAUGGUUUGAGUUUACAUCAUCGCAUGUAGUUUUACUCGCAUGUGUG ..((((((((((((.((((((((..(((((...)))))...)))))))).((.(((((((.((((((.....))))))..))))))))))))))))))))).....(((....))) ( -30.80) >consensus UCGACUAUAUGUGAAGCGAAUUCGAUUGGUAAAAUUAAACAGAAUUUGCCUGUAUGUGAAGCGAAUUCGAAUGGUUUGAGUUUACAUCAUCGCAUGUAGUUUUACUCGCAUGUGUG ..((((((((((((.((((((((..(((.......)))...)))))))).......(((.(..(((((((.....)))))))..).)))))))))))))))............... (-21.42 = -22.10 + 0.68)

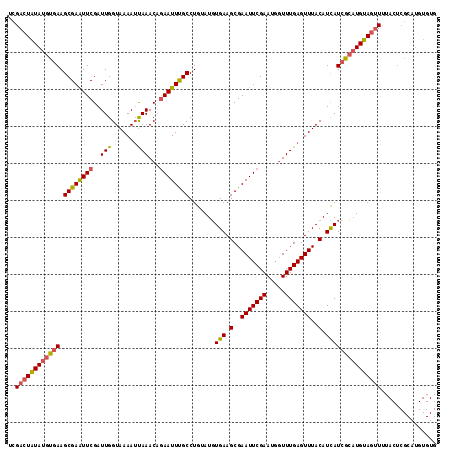
| Location | 1,996,754 – 1,996,867 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -20.34 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1996754 113 + 27905053 AAAGAUUCGAAUUCGCUUCAAAUUCAGGCGAAUUUGGUUUGUUUUUACCAAUCGAAUUCGCUUCACAUACAGUCGAAUUCGAUUUGAUUUUACUUUAUUACACGAGUCAAAUU ......(((((((((((........((((((((((((((.((....)).)))))))))))))).......)).)))))))))((((((((.............)))))))).. ( -31.98) >DroSec_CAF1 145894 112 + 1 AACCAUUCGAAUUCGCUUCACAUACAGGCAAAUUCUGUUUAAUUUUACCAAUCGAACUCGCUUCACAUAUAGUCGA-UUCGGUUUGAUUUUACAUUAUUACACGAGUUAAAUU ....(((((((((.((((.......)))).)))))(((..((((..(((((((((.((............))))))-)).)))..))))..))).........))))...... ( -20.10) >DroSim_CAF1 151098 113 + 1 AACCAUUCGAAUUCGCUUCACAUACAGGCAAAUUCUGUUUAAUUUUACCAAUCGAAUUCGCUUCACAUAUAGUCGAAUUCGGUUUUAUUUUACAUUAUUACACGAGUUAAAUU ....(((((((((.((((.......)))).)))))(((..(((......(((((((((((((........)).)))))))))))..)))..))).........))))...... ( -21.30) >consensus AACCAUUCGAAUUCGCUUCACAUACAGGCAAAUUCUGUUUAAUUUUACCAAUCGAAUUCGCUUCACAUAUAGUCGAAUUCGGUUUGAUUUUACAUUAUUACACGAGUUAAAUU ....(((((((((.((((.......)))).)))))(((..((((.....(((((((((((((........)).))))))))))).))))..))).........))))...... (-20.34 = -20.23 + -0.10)

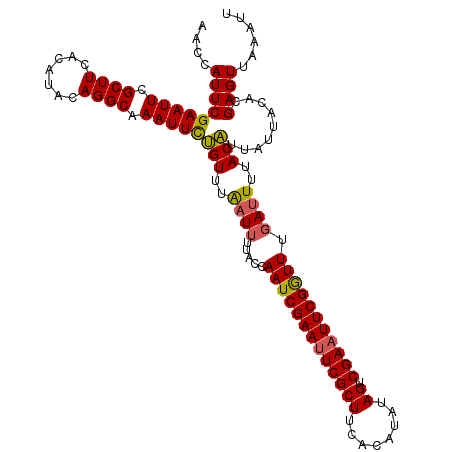
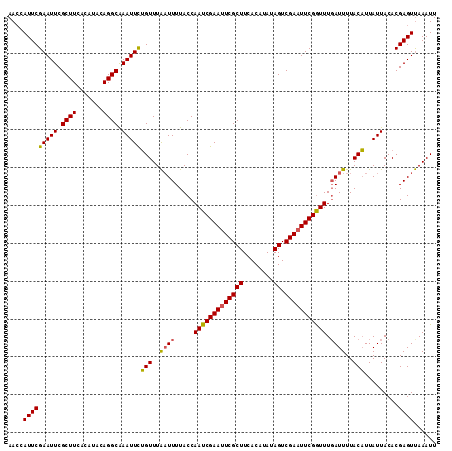
| Location | 1,996,794 – 1,996,903 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -15.60 |
| Consensus MFE | -14.49 |
| Energy contribution | -14.60 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1996794 109 + 27905053 GUUUUUACCAAUCGAAUUCGCUUCACAUACAGUCGAAUUCGAUUUGAUUUUACUUUAUUACACGAGUCAAAUUACGUGCAUAUAUAUA----GACCUCCUUCGACCAUAUAUA .........(((((((((((((........)).)))))))))))((((((.((((........)))).)))))).....((((((...----((......))....)))))). ( -18.10) >DroSec_CAF1 145934 102 + 1 AAUUUUACCAAUCGAACUCGCUUCACAUAUAGUCGA-UUCGGUUUGAUUUUACAUUAUUACACGAGUUAAAUUACGUGCGUACUUAUA--------UCCUUUGACUAU--UUA .........((((((((.((..((((.....)).))-..))))))))))...............(((((((..(..((......))..--------)..)))))))..--... ( -12.50) >DroSim_CAF1 151138 107 + 1 AAUUUUACCAAUCGAAUUCGCUUCACAUAUAGUCGAAUUCGGUUUUAUUUUACAUUAUUACACGAGUUAAAUUACGUGCGUACUUAUAUUUAUUUAUCCUUUGACU------A .........(((((((((((((........)).)))))))))))....................(((((((.((.(((.(((....))).))).))...)))))))------. ( -16.20) >consensus AAUUUUACCAAUCGAAUUCGCUUCACAUAUAGUCGAAUUCGGUUUGAUUUUACAUUAUUACACGAGUUAAAUUACGUGCGUACUUAUA________UCCUUUGACUAU___UA .........(((((((((((((........)).)))))))))))................((((..........))))................................... (-14.49 = -14.60 + 0.11)

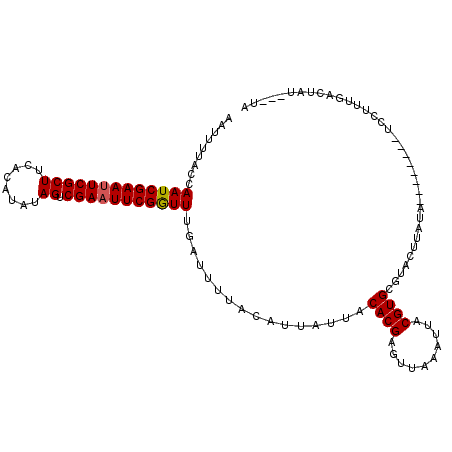
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:34 2006