| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,169,436 – 14,169,526 |
| Length | 90 |
| Max. P | 0.877989 |

| Location | 14,169,436 – 14,169,526 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -20.44 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14169436 90 + 27905053 AGGCGUUGGGAAAAGCCAAGCGAAAAGUUGGUGGAAAUGCCAAAUGCGAAACGAAGCGAAAUCGCCUCGGGGCCAAGCCUAUGCAAAUGU ..((((.(((....(((..(((.....(((((......))))).)))....(((.(((....))).))).)))....)))))))...... ( -27.40) >DroSec_CAF1 161016 90 + 1 AGGCGUUGGGAAAAGCCAAGCGAAAAGUUGGUGGAAAUGUCAAAUGCGAAACGAAGCGAAAUCGCCUCGGGGCCAAGCCUAUGCAAAUGC .((((((.......(((((.(.....))))))...))))))...((((...(((.(((....))).)))((((...)))).))))..... ( -24.70) >DroSim_CAF1 46229 90 + 1 AGGCGUUGGGAAAAGCCAAGCGAAAAGUUGGUGGAAAUGCCAAAUGCGAAACGAAGCGAAAUCGCCUCGGGGCCAAGCCUAUGCAAAUGC ..((((.(((....(((..(((.....(((((......))))).)))....(((.(((....))).))).)))....)))))))...... ( -27.40) >DroEre_CAF1 39237 90 + 1 AGGCGCAGGGAAAAGCCAAGCGAAAAGUUGCUGGAAAUGCCAAAUGCGAAACGAAGCGAAAUCGCCUCGGGGCCAAGCCUAUGCAAAUGU (((((((.((.....(((.((((....))))))).....))...)))....(((.(((....))).))).......)))).......... ( -27.90) >DroYak_CAF1 22907 90 + 1 AGGCGUAGGGAAAAACCGAGCGAGAAGUUGGUGGAAAUGGCAAAUGGGAAACGAAGCGAAAUCCCCUCGGGGCCAAGCCUAUGCAAAUGU ..(((((((.....(((.(((.....)))))).....((((.....(....)..........(((...)))))))..)))))))...... ( -25.80) >consensus AGGCGUUGGGAAAAGCCAAGCGAAAAGUUGGUGGAAAUGCCAAAUGCGAAACGAAGCGAAAUCGCCUCGGGGCCAAGCCUAUGCAAAUGU .((((((.......(((((.(.....))))))...))))))...((((...(((.(((....))).)))((((...)))).))))..... (-20.44 = -21.40 + 0.96)

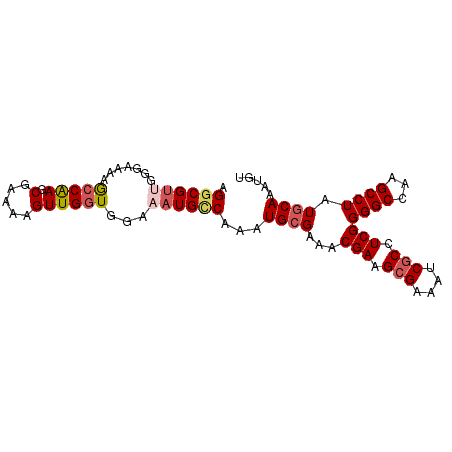

| Location | 14,169,436 – 14,169,526 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -22.55 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14169436 90 - 27905053 ACAUUUGCAUAGGCUUGGCCCCGAGGCGAUUUCGCUUCGUUUCGCAUUUGGCAUUUCCACCAACUUUUCGCUUGGCUUUUCCCAACGCCU .((..(((...(((...))).(((((((....)))))))....)))..))...................((((((......)))).)).. ( -23.30) >DroSec_CAF1 161016 90 - 1 GCAUUUGCAUAGGCUUGGCCCCGAGGCGAUUUCGCUUCGUUUCGCAUUUGACAUUUCCACCAACUUUUCGCUUGGCUUUUCCCAACGCCU ((....))..((((((((...(((((((....)))))))....................((((........))))......)))).)))) ( -23.00) >DroSim_CAF1 46229 90 - 1 GCAUUUGCAUAGGCUUGGCCCCGAGGCGAUUUCGCUUCGUUUCGCAUUUGGCAUUUCCACCAACUUUUCGCUUGGCUUUUCCCAACGCCU ((....))..((((((((...(((((((....)))))))....((....(((.................)))..)).....)))).)))) ( -24.13) >DroEre_CAF1 39237 90 - 1 ACAUUUGCAUAGGCUUGGCCCCGAGGCGAUUUCGCUUCGUUUCGCAUUUGGCAUUUCCAGCAACUUUUCGCUUGGCUUUUCCCUGCGCCU ......((.((((...((((.(((((((....)))))))....((.....))......(((........))).))))....)))).)).. ( -24.60) >DroYak_CAF1 22907 90 - 1 ACAUUUGCAUAGGCUUGGCCCCGAGGGGAUUUCGCUUCGUUUCCCAUUUGCCAUUUCCACCAACUUCUCGCUCGGUUUUUCCCUACGCCU ......((.((((...((..(((((((((...((...))..))))..(((..........))).......)))))..))..)))).)).. ( -17.70) >consensus ACAUUUGCAUAGGCUUGGCCCCGAGGCGAUUUCGCUUCGUUUCGCAUUUGGCAUUUCCACCAACUUUUCGCUUGGCUUUUCCCAACGCCU .((..(((...(((...))).(((((((....)))))))....)))..))...................((((((......)))).)).. (-18.84 = -19.48 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:38 2006