
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,166,388 – 14,166,501 |
| Length | 113 |
| Max. P | 0.953686 |

| Location | 14,166,388 – 14,166,501 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -16.90 |
| Energy contribution | -17.79 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
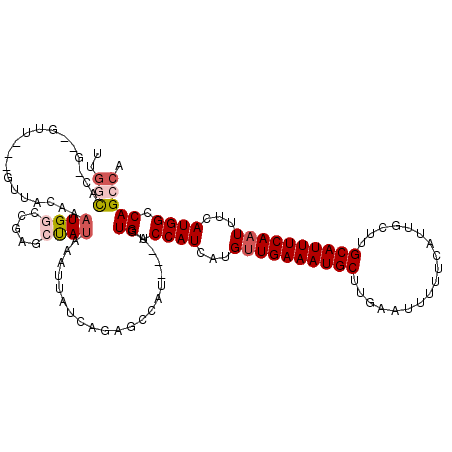
>3R_DroMel_CAF1 14166388 113 + 27905053 UUGGCACGGU-GUA---GUUACAAAUGGUCGGGCUAUAAAUUGCCACAGCCAU---AGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCUUGCAUUUCAAUUUCAUGGCCAGCCA .((((...((-(..---..)))..(((((.((((........))).).)))))---..((.((((...((((((((((..((((.....))).)..))))))))))...)))).)))))) ( -33.50) >DroVir_CAF1 42680 113 + 1 U---CAGAGC-GUUCAUUGUGGCAUUGCCUUGGCCAUAAAUUAAAGCCGCUCU---AGUGUCCAUCAUGUUGAAAUGCUUGCAUUUUUCAUAGCUUGCAUUUCAAUUUCAUGGCCAGCCA .---.(((((-(....(((((((.........)))))))........))))))---..((.((((...((((((((((..((..........))..))))))))))...)))).)).... ( -34.60) >DroPse_CAF1 46102 111 + 1 GUGGCA------GC---GUUACGAAUGGGCGAGCUAUAAAUUAUCAGAGCCAUAGUAGUGUCCAUCACGUUGAAAUGCUUGAAUUUUUCAUCGCUUGCAUUUCAAUUUCAUGGGCAGCCA .((((.------..---.((((.....(((...((..........)).)))...))))(((((((.(.((((((((((.(((........)))...)))))))))).).))))))))))) ( -31.90) >DroWil_CAF1 38237 114 + 1 UUGUUGCUGUCAUU---AUUAUAAAUGCUCAGACUAUAAAUUAUCGGCGCUCU---AGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCUUGCAUUUCAUUUUCAUGGCCAGCCA ..(((((((.((((---......))))..))).............(((((...---.)))))..(((((.((((((((..((((.....))).)..))))))))....))))).)))).. ( -23.10) >DroAna_CAF1 35021 112 + 1 UUGGCACAAC-GUU---UUUACAAAUGGCCGAGCUAUAAAUUGCCU-GCCCAU---AGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCAUGCAUUUCAAUUUCAUGGCCAGCCA .((((.....-...---.......((((.((.((........)).)-).))))---..((.((((...((((((((((.(((((.....))).)).))))))))))...)))).)))))) ( -29.80) >DroPer_CAF1 44303 111 + 1 GUGGCA------GC---GUUACGAAUGGGCGAGCUAUAAAUUAUCAGAGCCAUACUAGUGUCCAUCACGUUGAAAUGCUUGAAUUUUUCAUCGCUUGCAUUUCAAUUUCAUGGGCAGCCA .((((.------..---..........(((...((..........)).))).......(((((((.(.((((((((((.(((........)))...)))))))))).).))))))))))) ( -30.30) >consensus UUGGCAC_G__GUU___GUUACAAAUGGCCGAGCUAUAAAUUAUCAGAGCCAU___AGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCUUGCAUUUCAAUUUCAUGGCCAGCCA ..(((...................((((.....)))).....................((.((((...((((((((((..................))))))))))...)))).))))). (-16.90 = -17.79 + 0.89)


| Location | 14,166,388 – 14,166,501 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -18.17 |
| Energy contribution | -17.15 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14166388 113 - 27905053 UGGCUGGCCAUGAAAUUGAAAUGCAAGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACU---AUGGCUGUGGCAAUUUAUAGCCCGACCAUUUGUAAC---UAC-ACCGUGCCAA (((((((((((.(..(((((((((..(.(((.....))))..)))))))))..).))))(((..---((((.((.(((........))))).)))).)))...---...-.))).)))). ( -34.00) >DroVir_CAF1 42680 113 - 1 UGGCUGGCCAUGAAAUUGAAAUGCAAGCUAUGAAAAAUGCAAGCAUUUCAACAUGAUGGACACU---AGAGCGGCUUUAAUUUAUGGCCAAGGCAAUGCCACAAUGAAC-GCUCUG---A ....((.((((.(..(((((((((..((..........))..)))))))))..).)))).)).(---(((((((((.........))))..(((...))).........-))))))---. ( -34.50) >DroPse_CAF1 46102 111 - 1 UGGCUGCCCAUGAAAUUGAAAUGCAAGCGAUGAAAAAUUCAAGCAUUUCAACGUGAUGGACACUACUAUGGCUCUGAUAAUUUAUAGCUCGCCCAUUCGUAAC---GC------UGCCAC ((((.((((((.(..(((((((((....((........))..)))))))))..).))))..........((((.(((....))).))))..............---))------.)))). ( -27.50) >DroWil_CAF1 38237 114 - 1 UGGCUGGCCAUGAAAAUGAAAUGCAAGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACU---AGAGCGCCGAUAAUUUAUAGUCUGAGCAUUUAUAAU---AAUGACAGCAACAA ..((((.((((.(...((((((((..(.(((.....))))..))))))))...).)))).....---...((.(.(((........))).).)).........---.....))))..... ( -23.00) >DroAna_CAF1 35021 112 - 1 UGGCUGGCCAUGAAAUUGAAAUGCAUGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACU---AUGGGC-AGGCAAUUUAUAGCUCGGCCAUUUGUAAA---AAC-GUUGUGCCAA ((((...((((.(..(((((((((.((.(((.....))))).)))))))))..).))))(((..---((((.(-..((........))..).)))).)))...---...-.....)))). ( -33.70) >DroPer_CAF1 44303 111 - 1 UGGCUGCCCAUGAAAUUGAAAUGCAAGCGAUGAAAAAUUCAAGCAUUUCAACGUGAUGGACACUAGUAUGGCUCUGAUAAUUUAUAGCUCGCCCAUUCGUAAC---GC------UGCCAC ((((.((((((.(..(((((((((....((........))..)))))))))..).))))......(..(((((.(((....))).))).))..).........---))------.)))). ( -27.80) >consensus UGGCUGGCCAUGAAAUUGAAAUGCAAGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACU___AUGGCUCUGAUAAUUUAUAGCCCGACCAUUUGUAAC___AAC__C_GUGCCAA .(((((.((((.(..(((((((((..(.(((.....))))..)))))))))..).))))...((.....)).............)))))............................... (-18.17 = -17.15 + -1.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:35 2006