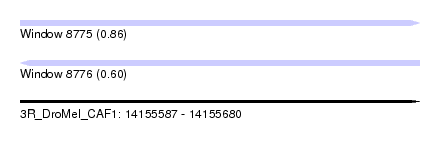
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,155,587 – 14,155,680 |
| Length | 93 |
| Max. P | 0.864013 |

| Location | 14,155,587 – 14,155,680 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 72.78 |
| Mean single sequence MFE | -18.41 |
| Consensus MFE | -5.84 |
| Energy contribution | -6.62 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
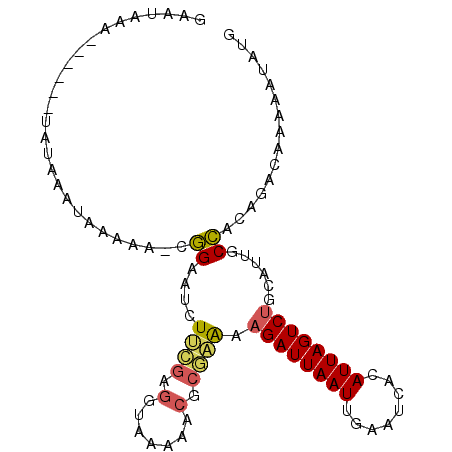
>3R_DroMel_CAF1 14155587 93 + 27905053 CAUAUUUUUGUCUGCGGCAAUGCAGACUAAUGUGAUUCAAUUAAUCUUUUCGCGUUUUUACCUCGAAGAUUCCU-UUUUUAUUUAUA------UUUAUUC ((((((...(((((((....))))))).))))))........(((((((..(.((....)))..)))))))...-............------....... ( -17.80) >DroSec_CAF1 148447 90 + 1 CAUAUUUUUGUCUGCGGCAAUGCAGACUAAUGUGAUUCAAUUAAUCUUUUCGCGUUUU---CGCGAAGAUUCCG-UUUUCAUUUAUA------UUUAUUC ((((((...(((((((....))))))).))))))............((((((((....---)))))))).....-............------....... ( -24.00) >DroSim_CAF1 33766 93 + 1 CAUAUUUUUGUCUGCGGCAAUGCAGACUAAUGUGAUUCAAUUAAUCUUUUCGCGUUUUUACCGCGAAGAUUCCG-UUUUCAUUUAUA------UUUAUUC ((((((...(((((((....))))))).))))))............((((((((.......)))))))).....-............------....... ( -23.20) >DroEre_CAF1 27519 87 + 1 CAUAUUUUUGCAUGUGGCAACGCAGACUAAUGUGGUUCAAUUAAUCUUUUCGCGUUUUUACCUCGAAGAUGCCU-UUUUU------A------UUUACUC ((((((...(..((((....))))..).)))))).................((((((((.....))))))))..-.....------.------....... ( -16.60) >DroAna_CAF1 25596 88 + 1 CAUAUUUUUGUAUGUGGAAACUCAGACUAAUGUAAUUCAAUUAAUCUUCCAGUGGGUUCUUCUCUUAAAUCCCCCCUUUU------A------UCCCUCC (((((....)))))((((.....(((.((((........)))).)))))))..(((..............))).......------.------....... ( -8.94) >DroPer_CAF1 30645 95 + 1 CAUAUUUUUGUGUGUAGAGUCGCCGACUAAUGUAAUUCAAUUAAUCUUCCGGCGUCGG--CGUUGGAG--UCUG-CUCUCUUCUCUCUCCUUCUCUCUUC (((((....))))).((((..((.(((((((........))))....(((((((....--))))))))--)).)-).))))................... ( -19.90) >consensus CAUAUUUUUGUCUGCGGCAACGCAGACUAAUGUGAUUCAAUUAAUCUUUUCGCGUUUUUACCUCGAAGAUUCCG_UUUUCAUUUAUA______UUUAUUC ((((((...((.((((....)))).)).))))))............((((((...........))))))............................... ( -5.84 = -6.62 + 0.78)



| Location | 14,155,587 – 14,155,680 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 72.78 |
| Mean single sequence MFE | -18.14 |
| Consensus MFE | -5.66 |
| Energy contribution | -5.55 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14155587 93 - 27905053 GAAUAAA------UAUAAAUAAAAA-AGGAAUCUUCGAGGUAAAAACGCGAAAAGAUUAAUUGAAUCACAUUAGUCUGCAUUGCCGCAGACAAAAAUAUG .......------...........(-(..((((((((.(.......).)))..)))))..))...........((((((......))))))......... ( -15.20) >DroSec_CAF1 148447 90 - 1 GAAUAAA------UAUAAAUGAAAA-CGGAAUCUUCGCG---AAAACGCGAAAAGAUUAAUUGAAUCACAUUAGUCUGCAUUGCCGCAGACAAAAAUAUG .......------....((((....-.......((((((---....))))))..((((.....)))).)))).((((((......))))))......... ( -20.50) >DroSim_CAF1 33766 93 - 1 GAAUAAA------UAUAAAUGAAAA-CGGAAUCUUCGCGGUAAAAACGCGAAAAGAUUAAUUGAAUCACAUUAGUCUGCAUUGCCGCAGACAAAAAUAUG .......------....((((....-.......((((((.......))))))..((((.....)))).)))).((((((......))))))......... ( -21.50) >DroEre_CAF1 27519 87 - 1 GAGUAAA------U------AAAAA-AGGCAUCUUCGAGGUAAAAACGCGAAAAGAUUAAUUGAACCACAUUAGUCUGCGUUGCCACAUGCAAAAAUAUG .......------.------.....-.((((((.....)))...(((((....((((((((........))))))))))))))))............... ( -14.80) >DroAna_CAF1 25596 88 - 1 GGAGGGA------U------AAAAGGGGGGAUUUAAGAGAAGAACCCACUGGAAGAUUAAUUGAAUUACAUUAGUCUGAGUUUCCACAUACAAAAAUAUG (((((..------.------...(((((...(((....)))...))).))...((((((((........))))))))...)))))............... ( -14.50) >DroPer_CAF1 30645 95 - 1 GAAGAGAGAAGGAGAGAGAAGAGAG-CAGA--CUCCAACG--CCGACGCCGGAAGAUUAAUUGAAUUACAUUAGUCGGCGACUCUACACACAAAAAUAUG ....((((..((((...........-....--))))..((--(((((..(....)..((((........))))))))))).))))............... ( -22.36) >consensus GAAUAAA______UAUAAAUAAAAA_CGGAAUCUUCGAGGUAAAAACGCGAAAAGAUUAAUUGAAUCACAUUAGUCUGCAUUGCCACAGACAAAAAUAUG ...........................((....((((.(.......).)))).((((((((........))))))))......))............... ( -5.66 = -5.55 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:30 2006