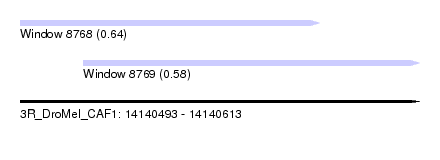
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,140,493 – 14,140,613 |
| Length | 120 |
| Max. P | 0.643735 |

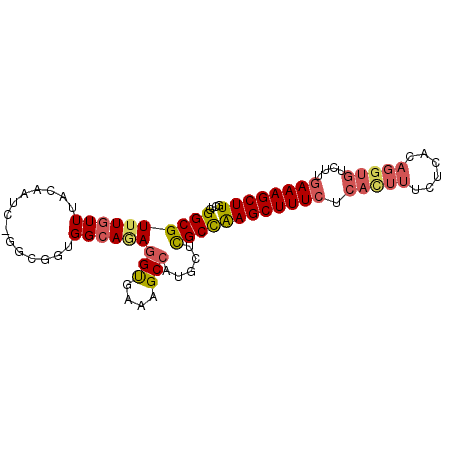
| Location | 14,140,493 – 14,140,583 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 84.90 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -18.62 |
| Energy contribution | -20.66 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14140493 90 + 27905053 GCUGGGCGUUUGUUUACAAUCCGGCUGUGGCAGAGGUGAAAGCCAUGCUCGCCAAGCUUUCUCAUUUUCUCACAGGUGUCUUGAAAGCUU ((((((...(((....))))))))).((((((..(((....))).))).))).((((((((.(((((......)))))....)))))))) ( -28.80) >DroSec_CAF1 12701 89 + 1 GCUGGGCGUUUGUUUACAAUC-GGCGGUGGCAAAAGCGAAAGCCAUGCUCGCCAAGCUUUCUCACUUUCUCACAGGUGUCUUGAAAGCUU (((((..((......))..))-)))(((((((...((....))..))).))))((((((((.(((((......)))))....)))))))) ( -33.10) >DroSim_CAF1 13006 89 + 1 GCUGGGCGUUUGUUUACAAUC-GGCGGUGGCAGAGGCGAAAGCCAUGCUCGCCAAGCUUUCUCACUUUCUCACAGGUGUCUUGAAAGCUU (((((..((......))..))-)))(((((((..(((....))).))).))))((((((((.(((((......)))))....)))))))) ( -37.60) >DroEre_CAF1 13236 89 + 1 GCUGGGCGUUUGUUUACAAUC-GGCGGUGGAAGAGGUGAAAGCCAUGCUCGCCAAGCUUUCUCACUUUCUCACAGGUGUCUUGAAAGCUU (((((..((......))..))-)))(((((..(.(((....))).)..)))))((((((((.(((((......)))))....)))))))) ( -29.90) >DroAna_CAF1 12502 77 + 1 UUUGGGCGUUUGUUUACAU-----CAGAGGCAUAGGUGAAAGCCGCUGUUGCUGAGCUUU--------CUCACAGGUGUUUUAAAAGCUU ...((((.((((...((((-----(((..(((..(((....)))..)))..))(((....--------)))...)))))..)))).)))) ( -19.50) >consensus GCUGGGCGUUUGUUUACAAUC_GGCGGUGGCAGAGGUGAAAGCCAUGCUCGCCAAGCUUUCUCACUUUCUCACAGGUGUCUUGAAAGCUU ....((((((((((..............))))))(((....))).....))))((((((((.(((((......)))))....)))))))) (-18.62 = -20.66 + 2.04)

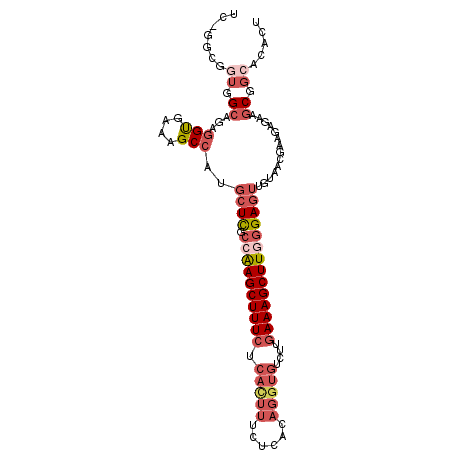
| Location | 14,140,512 – 14,140,613 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.88 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -20.82 |
| Energy contribution | -24.54 |
| Covariance contribution | 3.72 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14140512 101 + 27905053 UCCGGCUGUGGCAGAGGUGAAAGCCAUGCUCGCCAAGCUUUCUCAUUUUCUCACAGGUGUCUUGAAAGCUUGGGAGUUGUAACGAAGAGAAGCGGCCCACU ...((((((..(...(((....)))..((((.((((((((((.(((((......)))))....))))))))))))))...........)..)))))).... ( -38.10) >DroSec_CAF1 12720 100 + 1 UC-GGCGGUGGCAAAAGCGAAAGCCAUGCUCGCCAAGCUUUCUCACUUUCUCACAGGUGUCUUGAAAGCUUGGGAGUUGUAACGAAGAGAAGCGGCACACU ..-(((..(.((....)).)..)))..((((.((((((((((.(((((......)))))....))))))))))))))(((..((........))..))).. ( -35.90) >DroSim_CAF1 13025 100 + 1 UC-GGCGGUGGCAGAGGCGAAAGCCAUGCUCGCCAAGCUUUCUCACUUUCUCACAGGUGUCUUGAAAGCUUGGGAGUUGUAACGAAGAGAAGCGGCACACU ..-.((.((..(...(((....)))..((((.((((((((((.(((((......)))))....))))))))))))))...........)..)).))..... ( -39.00) >DroEre_CAF1 13255 100 + 1 UC-GGCGGUGGAAGAGGUGAAAGCCAUGCUCGCCAAGCUUUCUCACUUUCUCACAGGUGUCUUGAAAGCUUCGGAGUUGUAACGAAGAGAAACGGACCACU ..-...(((((....(((....)))..((((.(.((((((((.(((((......)))))....)))))))).)))))...................))))) ( -29.10) >DroAna_CAF1 12521 88 + 1 -----CAGAGGCAUAGGUGAAAGCCGCUGUUGCUGAGCUUU--------CUCACAGGUGUUUUAAAAGCUUCCGAGUUUUAAUACGGAGAGGCAAGUGAAU -----(((..(((..(((....)))..)))..))).((((.--------(((....(((((...((((((....))))))))))).)))))))........ ( -23.60) >consensus UC_GGCGGUGGCAGAGGUGAAAGCCAUGCUCGCCAAGCUUUCUCACUUUCUCACAGGUGUCUUGAAAGCUUGGGAGUUGUAACGAAGAGAAGCGGCACACU .......((.((...(((....)))..((((.((((((((((.(((((......)))))....))))))))))))))..............)).))..... (-20.82 = -24.54 + 3.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:24 2006