| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,122,511 – 14,122,610 |
| Length | 99 |
| Max. P | 0.774953 |

| Location | 14,122,511 – 14,122,610 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14122511 99 + 27905053 UGCGGUCUGAAAAUUGCACGUUUGCAUGUAAAAUAUAUGGGGAGCAGCAGAUGAAAUUCAUCACUGAA-------UG---CUCCUCCAUCUCCUAC---CCCUGCAAUCCAC (((((........(((((.(....).))))).....((((((((((.(((.(((......))))))..-------))---)))).)))).......---..)))))...... ( -26.80) >DroSec_CAF1 9711 109 + 1 UGCGGUCUGAAAAUUGCACGUUUGCAUGUAAAAUAUAUGGGGAGCAGCAGAUGAAAUCCAUCACUGAAAUGUGGAUGCUCCUCCUCCAUCUCCUAC---GCCCGCAAUCCAC (((((.(.......((((....)))).(((......((((((((.((((((((.....))))((......))...)))).)))).))))....)))---).)))))...... ( -29.80) >DroEre_CAF1 9380 97 + 1 UGCGAGCUGAAAAUUGCACGUUUGCAUGUAAAAUAUACGAGGAGCAGCAGAUGAAAUCCAUCACUGAAAUGUGGAGACUCCUCC---------------CCUUGCAAUCCAC (((((((((.......)).)))))))...........(((((.(((.(((.(((......))))))...)))((((....))))---------------)))))........ ( -23.40) >DroYak_CAF1 12654 112 + 1 UGCGAGCUGAAAAUUGCACGUUUGCAUGUAAAAUAUACGAGGAGCAGCAGAUGAAAUCCAUCGCUGAAAUGUGGGGACUCCUCCUCCGUCUUCUGCUCCGCUUGCAAUCCAC (((((((......(((((.(....).))))).........(((((((.(((((.........((......))(((((....)))))))))).))))))))))))))...... ( -38.10) >consensus UGCGAGCUGAAAAUUGCACGUUUGCAUGUAAAAUAUACGAGGAGCAGCAGAUGAAAUCCAUCACUGAAAUGUGGAGACUCCUCCUCCAUCUCCUAC___CCCUGCAAUCCAC (((((((......(((((.(....).))))).......(((((((((..((((.....)))).)))......(....)..)))))).............)))))))...... (-19.12 = -19.88 + 0.75)


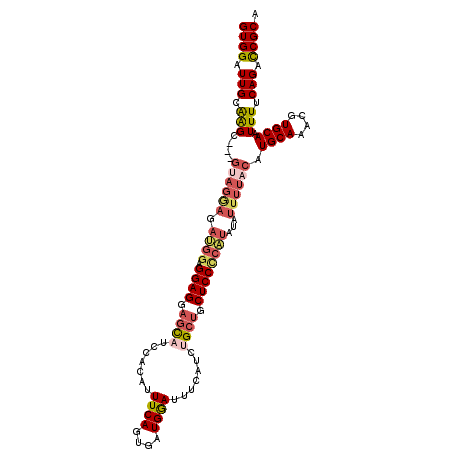
| Location | 14,122,511 – 14,122,610 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -22.72 |
| Energy contribution | -24.84 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14122511 99 - 27905053 GUGGAUUGCAGGG---GUAGGAGAUGGAGGAG---CA-------UUCAGUGAUGAAUUUCAUCUGCUGCUCCCCAUAUAUUUUACAUGCAAACGUGCAAUUUUCAGACCGCA ((((((((((.(.---((((((.((((.((((---((-------..((((((......))).))).))))))))))...)))))).......).)))))).......)))). ( -34.41) >DroSec_CAF1 9711 109 - 1 GUGGAUUGCGGGC---GUAGGAGAUGGAGGAGGAGCAUCCACAUUUCAGUGAUGGAUUUCAUCUGCUGCUCCCCAUAUAUUUUACAUGCAAACGUGCAAUUUUCAGACCGCA ......(((((..---((((((.((((.((((.(((((((((((....))).)))).......)))).))))))))...)))))).((((....)))).........))))) ( -38.51) >DroEre_CAF1 9380 97 - 1 GUGGAUUGCAAGG---------------GGAGGAGUCUCCACAUUUCAGUGAUGGAUUUCAUCUGCUGCUCCUCGUAUAUUUUACAUGCAAACGUGCAAUUUUCAGCUCGCA ((((.(((((.((---------------((((.(((........((((....))))........))).))))))(((.....))).)))))....((........)))))). ( -26.59) >DroYak_CAF1 12654 112 - 1 GUGGAUUGCAAGCGGAGCAGAAGACGGAGGAGGAGUCCCCACAUUUCAGCGAUGGAUUUCAUCUGCUGCUCCUCGUAUAUUUUACAUGCAAACGUGCAAUUUUCAGCUCGCA ...........(((.(((.(((((..((((((..((....))....((((((((.....)))).))))))))))(((.....))).((((....)))).))))).)))))). ( -32.50) >consensus GUGGAUUGCAAGC___GUAGGAGAUGGAGGAGGAGCAUCCACAUUUCAGUGAUGGAUUUCAUCUGCUGCUCCCCAUAUAUUUUACAUGCAAACGUGCAAUUUUCAGACCGCA ((((.(((.(((....((((((.((((.((((.((((.......((((....)))).......)))).))))))))...)))))).((((....)))).))).))).)))). (-22.72 = -24.84 + 2.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:20 2006