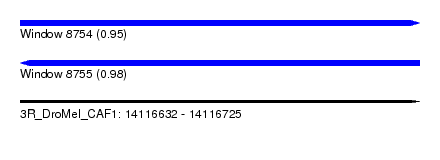
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,116,632 – 14,116,725 |
| Length | 93 |
| Max. P | 0.977292 |

| Location | 14,116,632 – 14,116,725 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -47.78 |
| Consensus MFE | -39.73 |
| Energy contribution | -41.20 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
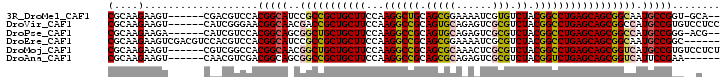
>3R_DroMel_CAF1 14116632 93 + 27905053 --UGC-ACCGGCAUUGCCGCUGCUCAGGCCGUAGACACGAUUUUCCGCUGCAGCCUUGGAAGCAGCGGCGGAUGCCGUGGACGUCG------ACUUCUUGCG --.((-(..((((((((((((((((((((.((((...((......)))))).))))..).))))))))).))))))((.(....).------))....))). ( -41.60) >DroVir_CAF1 1401 96 + 1 GGAGGACACGGCAUGGCCGCUGCUCAGGCCGUAGACGCGACUCUGCACUGCGGCCUUGGAAGCAGCGGUCGUUGCCGUUCCCGAUG------ACUUCUUGCG .(.(((.(((((((((((((((((((((((((((..(((....))).)))))))))..).))))))))))).))))))))))....------.......... ( -52.10) >DroPse_CAF1 5505 93 + 1 --CGU-CCCGGCAUGGCCGCUGCUCAGGCCGUAGACGCGACUCUGCACUGCGGCCUUGGAAGCAGCAGCCGCUGCCGUGGACGAUG------UCUUCUUGCG --(((-(((((((((((.((((((((((((((((..(((....))).)))))))))..).)))))).)))).))))).)))))...------.......... ( -53.40) >DroEre_CAF1 3835 96 + 1 ------GCCGGCAUUGCCGCUGCUCAGGCCGUAGACGCGAUUUUCCGCUGCGGCCUUGGAAGCAGCGGCGGAUGCCGUGGACGUGGACGUCGACUUCUUGCG ------((.(((((((((((((((((((((((((.((.(.....))))))))))))..).))))))))).))))))((.((((....)))).)).....)). ( -55.70) >DroMoj_CAF1 1374 96 + 1 AGAGGACACGGCAUGACCGCUGCUCAGGCCGUAGACGCGAGUUUGCGCUGCGGCCUUGGAAGCAGCAGCCGUUGCCGUGGCCGACG------ACUUCUUGCG ...((.(((((((((.(.((((((((((((((((.((((....)))))))))))))..).)))))).).)).))))))).))....------.......... ( -48.50) >DroAna_CAF1 3991 90 + 1 ------UUCGGAAUGACCGCUGCUCAGACCGUAGACGCGACUCUGCGCUGCGGCCUUGGAAGCAGCGGCCGCUGCCGUCGACGUUG------ACUUCUUGCG ------...((..((.(((((((((((.((((((.((((....)))))))))).))..).)))))))).))...))((((....))------))........ ( -35.40) >consensus ___G__CCCGGCAUGGCCGCUGCUCAGGCCGUAGACGCGACUCUGCGCUGCGGCCUUGGAAGCAGCGGCCGAUGCCGUGGACGAUG______ACUUCUUGCG ........((((((((((((((((((((((((((.((((....)))))))))))))..).))))))))))).)))))......................... (-39.73 = -41.20 + 1.47)



| Location | 14,116,632 – 14,116,725 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -48.05 |
| Consensus MFE | -41.74 |
| Energy contribution | -41.72 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14116632 93 - 27905053 CGCAAGAAGU------CGACGUCCACGGCAUCCGCCGCUGCUUCCAAGGCUGCAGCGGAAAAUCGUGUCUACGGCCUGAGCAGCGGCAAUGCCGGU-GCA-- (....)....------....(..(.((((((..((((((((((...((((((..(((......))).....)))))))))))))))).))))))).-.).-- ( -42.90) >DroVir_CAF1 1401 96 - 1 CGCAAGAAGU------CAUCGGGAACGGCAACGACCGCUGCUUCCAAGGCCGCAGUGCAGAGUCGCGUCUACGGCCUGAGCAGCGGCCAUGCCGUGUCCUCC (....)....------....((((((((((..(.(((((((((...((((((.(((((......))).)).))))))))))))))))..)))))).)))).. ( -46.70) >DroPse_CAF1 5505 93 - 1 CGCAAGAAGA------CAUCGUCCACGGCAGCGGCUGCUGCUUCCAAGGCCGCAGUGCAGAGUCGCGUCUACGGCCUGAGCAGCGGCCAUGCCGGG-ACG-- (....)....------...(((((.(((((..(((((((((((...((((((.(((((......))).)).))))))))))))))))).)))))))-)))-- ( -55.90) >DroEre_CAF1 3835 96 - 1 CGCAAGAAGUCGACGUCCACGUCCACGGCAUCCGCCGCUGCUUCCAAGGCCGCAGCGGAAAAUCGCGUCUACGGCCUGAGCAGCGGCAAUGCCGGC------ .((........((((....))))..((((((..((((((((((...((((((.(((((.....).)).)).)))))))))))))))).))))))))------ ( -49.60) >DroMoj_CAF1 1374 96 - 1 CGCAAGAAGU------CGUCGGCCACGGCAACGGCUGCUGCUUCCAAGGCCGCAGCGCAAACUCGCGUCUACGGCCUGAGCAGCGGUCAUGCCGUGUCCUCU (....)....------....((.(((((((..(((((((((((...((((((.(((((......))).)).))))))))))))))))).))))))).))... ( -52.30) >DroAna_CAF1 3991 90 - 1 CGCAAGAAGU------CAACGUCGACGGCAGCGGCCGCUGCUUCCAAGGCCGCAGCGCAGAGUCGCGUCUACGGUCUGAGCAGCGGUCAUUCCGAA------ (....)..((------(......)))((.((.(((((((((((...((((((.(((((......))).)).))))))))))))))))).))))...------ ( -40.90) >consensus CGCAAGAAGU______CAACGUCCACGGCAACGGCCGCUGCUUCCAAGGCCGCAGCGCAAAGUCGCGUCUACGGCCUGAGCAGCGGCCAUGCCGGG__C___ (....)...................(((((..(((((((((((...((((((.(((((......))).)).))))))))))))))))).)))))........ (-41.74 = -41.72 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:10 2006