| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
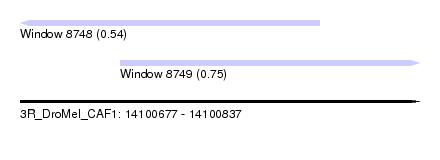
| Location | 14,100,677 – 14,100,837 |
| Length | 160 |
| Max. P | 0.753403 |

| Location | 14,100,677 – 14,100,797 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -27.71 |
| Consensus MFE | -22.85 |
| Energy contribution | -22.02 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
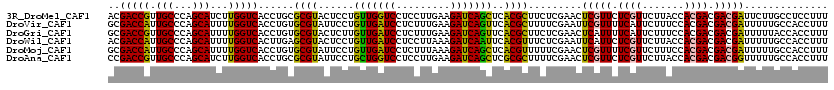
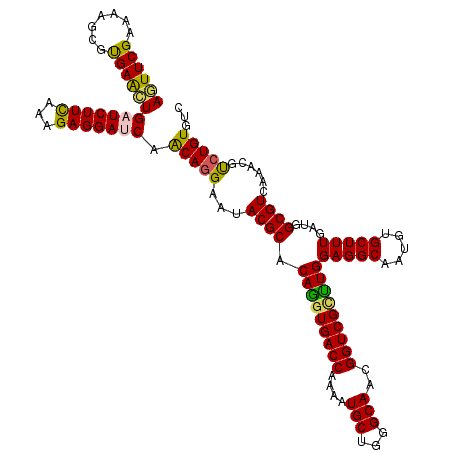
>3R_DroMel_CAF1 14100677 120 - 27905053 ACGACCGUUGCCCAGCAUCUUGGUCACCUGCGCGUACUCCUGUUGGUCCUCCUUGAAGAUCAGCUCACGCUUCUCGAACUCGUUCUCGUUCUUACCACGACGACGAUUCUUGCCUCCUUU ..(((((.(((...)))...)))))....((((((......(((((((.((...)).)))))))..)))).....(((.(((((.((((.......)))).))))))))..))....... ( -30.90) >DroVir_CAF1 23 120 - 1 GCGACCAUUGCCCAGCAUUUUGGUCACCUGUGCGUAUUCCUGUUGAUCCUCUUUGAAGAUCAGUUCACGCUUUUCGAAUUCGUUUUCAUUCUUUCCACGACGACGAUUUUUGCCACCUUU ..(((((.(((...)))...)))))....(((.(((.((.(((((........(((((((.(((((.........))))).))))))).........)))))..))....)))))).... ( -23.43) >DroGri_CAF1 234 120 - 1 GCGACCGUUGCCCAGCAUUUUGGUCACCUGUGCGUACUCUUGUUGAUCCUCUUUGAAGAUCAGUUCACGCUUCUCGAACUCAUUUUCAUUCUUUCCACGACGACGAUUUUUACCACCUUU ..(((((.(((...)))...)))))....(((.(((.((((((((........(((((((.(((((.........))))).))))))).........)))))).))....)))))).... ( -25.93) >DroWil_CAF1 19 120 - 1 ACGACCAUUGCCCAGCAUUUUGGUCACUUGAGCGUACUCCUGUUGAUCCUCCUUAAAGAUCAAUUCACGUUUCUCGAAUUCAUUCUCGUUCUUACCACGACGACGAUUUUUGCCACCUUU ..(((((.(((...)))...)))))..(((((...((....(((((((.........)))))))....))..)))))......(((((((........))))).)).............. ( -23.00) >DroMoj_CAF1 238 120 - 1 GCGACCAUUGCCCAGCAUUUUGGUCACCUGUGCGUAUUCCUGUUGAUCCUCUUUAAAGAUCAGCUCACGUUUUUCGAACUCGUUUUCGUUCUUUCCACGACGACGAUUUUUGCCACCUUU ..(((((.(((...)))...)))))....(((.(((.....(((((((.........)))))))...........(((.(((((.((((.......)))).))))).))))))))).... ( -28.00) >DroAna_CAF1 142 120 - 1 CCGACCGUUGCCCAGCAUCUUGGUCACCUGCGCGUAUUCCUGCUGGUCCUCCUUGAAGAUCAGCUCGCGCUUUUCGAACUCGUUCUCGUUCUUACCACGACGACGGUUUUUGCCACCUUU ..(((((.(((...)))...)))))....(((((.......(((((((.((...)).))))))).))))).....((((.((((.((((.......)))).))))))))........... ( -35.00) >consensus GCGACCAUUGCCCAGCAUUUUGGUCACCUGUGCGUACUCCUGUUGAUCCUCCUUGAAGAUCAGCUCACGCUUCUCGAACUCGUUCUCGUUCUUACCACGACGACGAUUUUUGCCACCUUU ..(((((.(((...)))...)))))......((((......(((((((.........)))))))..)))).........(((((.((((.......)))).))))).............. (-22.85 = -22.02 + -0.83)
| Location | 14,100,717 – 14,100,837 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -38.15 |
| Consensus MFE | -35.42 |
| Energy contribution | -34.53 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
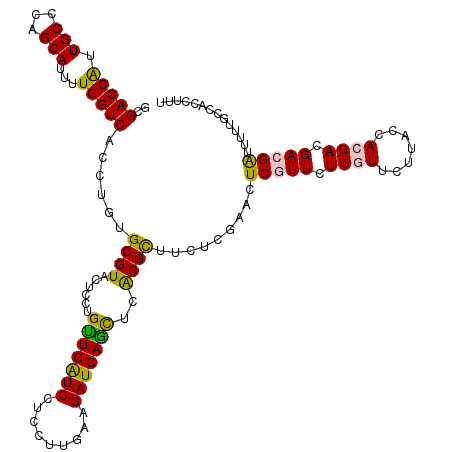
>3R_DroMel_CAF1 14100717 120 + 27905053 AGUUCGAGAAGCGUGAGCUGAUCUUCAAGGAGGACCAACAGGAGUACGCGCAGGUGACCAAGAUGCUGGGCAACGGUCGUCUGGAGGCAAUGUGCUUUGAUGGCGUCAAACGCCUGUGUC .((((((..(((....)))..))((....))))))..(((((.((((((.(((..((((....(((...)))..))))..)))(((((.....)))))....))))...)).)))))... ( -39.30) >DroVir_CAF1 63 120 + 1 AAUUCGAAAAGCGUGAACUGAUCUUCAAAGAGGAUCAACAGGAAUACGCACAGGUGACCAAAAUGCUGGGCAAUGGUCGCUUGGAGGCAAUGUGCUUUGAUGGCGUCAAACGUCUGUGUC ..................((((((((...))))))))(((((...((((.((((((((((...(((...))).))))))))))(((((.....)))))....))))......)))))... ( -37.50) >DroGri_CAF1 274 120 + 1 AGUUCGAGAAGCGUGAACUGAUCUUCAAAGAGGAUCAACAAGAGUACGCACAGGUGACCAAAAUGCUGGGCAACGGUCGCUUGGAGGCAAUGUGCUUUGAUGGCGUCAAACGUCUGUGUC ((((((.......))))))(((((((...))))))).((((((((((((.(((((((((....(((...)))..)))))))))...))...)))))))...((((.....)))))))... ( -37.70) >DroWil_CAF1 59 120 + 1 AAUUCGAGAAACGUGAAUUGAUCUUUAAGGAGGAUCAACAGGAGUACGCUCAAGUGACCAAAAUGCUGGGCAAUGGUCGUUUGGAGGCAAUGUGCUUUGAUGGCGUCAAACGUCUGUGUC ((((((.......))))))(((((((...))))))).((((..(((((((((((((((((...(((...))).))))))))))(((((.....)))))...)))))...))..))))... ( -33.20) >DroMoj_CAF1 278 120 + 1 AGUUCGAAAAACGUGAGCUGAUCUUUAAAGAGGAUCAACAGGAAUACGCACAGGUGACCAAAAUGCUGGGCAAUGGUCGCCUGGAGGCAAUGUGCUUUGAUGGCGUCAAACGUCUGUGUC ((((((.......))))))(((((((...))))))).(((((...((((.((((((((((...(((...))).))))))))))(((((.....)))))....))))......)))))... ( -41.40) >DroAna_CAF1 182 120 + 1 AGUUCGAAAAGCGCGAGCUGAUCUUCAAGGAGGACCAGCAGGAAUACGCGCAGGUGACCAAGAUGCUGGGCAACGGUCGGUUGGAGGCAAUGUGCUUUGAUGGCGUCAAGCGCCUGUGUC ...(((.......)))((((.(((((...))))).))))........(((((((((.....((((((...((((.....))))(((((.....)))))...))))))...))))))))). ( -39.80) >consensus AGUUCGAAAAGCGUGAACUGAUCUUCAAAGAGGAUCAACAGGAAUACGCACAGGUGACCAAAAUGCUGGGCAACGGUCGCUUGGAGGCAAUGUGCUUUGAUGGCGUCAAACGUCUGUGUC ((((((.......))))))(((((((...))))))).(((((...((((.(((((((((....(((...)))..)))))))))(((((.....)))))....))))......)))))... (-35.42 = -34.53 + -0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:04 2006