
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,078,850 – 14,078,956 |
| Length | 106 |
| Max. P | 0.849992 |

| Location | 14,078,850 – 14,078,956 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.52 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.43 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14078850 106 + 27905053 GCCUUGGCCUCCUCGGGCGCCUCCGGACACAAAUUACCCAGUGCCGCCAGUUCGAAUUUGUGCAGCUUUUUCUGCAUCAGCAAGCUGAA--U--UUGAA-AUAAU---UGA-AUA ...(((((......(((....)))((.(((..........))))))))))((((((((.((((((......))))))(((....)))))--)--)))))-.....---...-... ( -24.70) >DroVir_CAF1 2015 107 + 1 GCCUUGGCCUCCUCCGGCGCCUCCGGGCACAGAUUACCCAGUGCAGCCAGCUCGAACUUGUGCAGUUUCUUUUGCAUAAGCAAACUGGG--A--GUACACAAAAU---AAA-AUA ((((.(((((.....)).)))...))))........((((((((.....)).....(((((((((......)))))))))...))))))--.--...........---...-... ( -31.80) >DroGri_CAF1 1864 108 + 1 GCUUUGGCUUCCUCCGGCGCCUCUGGGCACAGAUUGCCCAGGGCAGCUAGUUCGAAUUUGUGCAGCUUCUUUUGCAUUAGCAGACUGGG--A--AUAAUUCAAAU---AAAUAUA ..((((..((..(((((((((.(((((((.....)))))))))).)))(((..(.....((((((......))))))...)..))).))--)--..))..)))).---....... ( -34.40) >DroWil_CAF1 1794 106 + 1 GCCUUGGCCUCUUCUGGAGCCUCCGGACACAGAUUGCCCAGUGCAGCCAAUUCGAAUUUGUGCAAUUUCUUUUGCAUUAGUAAACUGCAAAAAUAUGGA-AC--U---UU---UA ((((.(((.((((((((.....)))))...)))..))).)).))......((((.((((.((((.(((((........)).))).)))).)))).))))-..--.---..---.. ( -23.20) >DroMoj_CAF1 1901 104 + 1 GCCUUAGCCUCUUCCGGCGCCUCCGGGCAGAGAUUGCCCAGAGCGGCAAGUUCGAAUUUGUGCAGUUUCUUUUGCAUAAGUAAACUAGA--A--ACAAG-AAAAU---UGA---A (((...(((......)))((.((.((((((...)))))).))))))).((((...((((((((((......)))))))))).))))...--.--.(((.-....)---)).---. ( -33.00) >DroAna_CAF1 1602 104 + 1 GCCUUGGCUUCUUCCGGAGCCUCUGGGCACAGAUUGCCUAGGGCGGCUAGCUCGAAUUUAUGCAAUUUCUUUUGCAUCAGCAAGCUGGA---------A-AAAAAUCAUUA-CUA (((..((((((....)))))).(((((((.....))))))))))..((((((.(.....((((((......))))))...).)))))).---------.-...........-... ( -36.90) >consensus GCCUUGGCCUCCUCCGGCGCCUCCGGGCACAGAUUGCCCAGUGCAGCCAGUUCGAAUUUGUGCAGUUUCUUUUGCAUAAGCAAACUGGA__A__AUAAA_AAAAU___UAA_AUA ((((.(((.((....)).)))...(((((.....))))))).))..((((((.(.....((((((......))))))...).))))))........................... (-18.34 = -18.43 + 0.09)

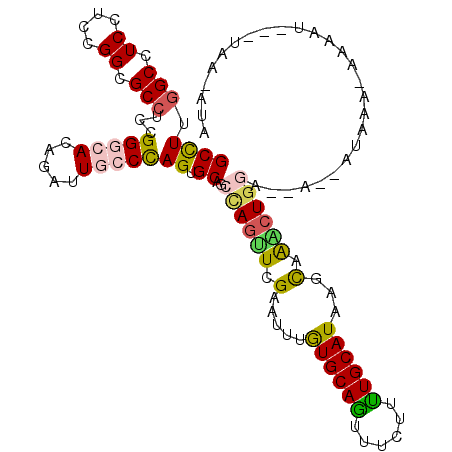

| Location | 14,078,850 – 14,078,956 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.52 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -21.38 |
| Energy contribution | -20.88 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14078850 106 - 27905053 UAU-UCA---AUUAU-UUCAA--A--UUCAGCUUGCUGAUGCAGAAAAAGCUGCACAAAUUCGAACUGGCGGCACUGGGUAAUUUGUGUCCGGAGGCGCCCGAGGAGGCCAAGGC ...-...---.....-.....--.--....((((.((..(((((......)))))..........(.((((.(.((((..(.....)..)))).).)))).))).))))...... ( -27.80) >DroVir_CAF1 2015 107 - 1 UAU-UUU---AUUUUGUGUAC--U--CCCAGUUUGCUUAUGCAAAAGAAACUGCACAAGUUCGAGCUGGCUGCACUGGGUAAUCUGUGCCCGGAGGCGCCGGAGGAGGCCAAGGC ...-...---.(((((.((.(--(--((.(((((((((.((((........)))).))))..)))))(((.((.(((((((.....)))))))..)))))))))...))))))). ( -36.60) >DroGri_CAF1 1864 108 - 1 UAUAUUU---AUUUGAAUUAU--U--CCCAGUCUGCUAAUGCAAAAGAAGCUGCACAAAUUCGAACUAGCUGCCCUGGGCAAUCUGUGCCCAGAGGCGCCGGAGGAAGCCAAAGC .......---.((((((((..--.--..((((.(((....)))......))))....))))))))(..((.((((((((((.....))))))).)))))..)............. ( -30.40) >DroWil_CAF1 1794 106 - 1 UA---AA---A--GU-UCCAUAUUUUUGCAGUUUACUAAUGCAAAAGAAAUUGCACAAAUUCGAAUUGGCUGCACUGGGCAAUCUGUGUCCGGAGGCUCCAGAAGAGGCCAAGGC ..---..---(--((-((...((((.((((((((.((........)))))))))).))))..)))))((((...((((((..((((....)))).)).))))....))))..... ( -29.10) >DroMoj_CAF1 1901 104 - 1 U---UCA---AUUUU-CUUGU--U--UCUAGUUUACUUAUGCAAAAGAAACUGCACAAAUUCGAACUUGCCGCUCUGGGCAAUCUCUGCCCGGAGGCGCCGGAAGAGGCUAAGGC .---...---.....-.((((--.--..((((((.(((......))))))))).))))..........(((((((((((((.....)))))))).))(((......)))...))) ( -30.90) >DroAna_CAF1 1602 104 - 1 UAG-UAAUGAUUUUU-U---------UCCAGCUUGCUGAUGCAAAAGAAAUUGCAUAAAUUCGAGCUAGCCGCCCUAGGCAAUCUGUGCCCAGAGGCUCCGGAAGAAGCCAAGGC ...-........(((-(---------(((((((((...((((((......)))))).....))))))....(((((.((((.....)))).)).)))...)))))))((....)) ( -32.80) >consensus UAU_UAA___AUUUU_UUUAU__U__UCCAGUUUGCUAAUGCAAAAGAAACUGCACAAAUUCGAACUGGCUGCACUGGGCAAUCUGUGCCCGGAGGCGCCGGAAGAGGCCAAGGC .............................((((((....((((........))))......)))))).(((...(((((((.....))))))).(((..(....)..)))..))) (-21.38 = -20.88 + -0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:55 2006