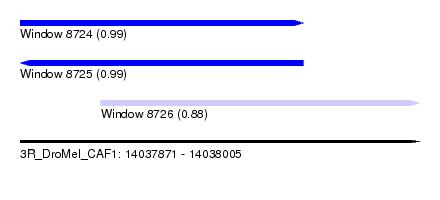
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,037,871 – 14,038,005 |
| Length | 134 |
| Max. P | 0.988731 |

| Location | 14,037,871 – 14,037,966 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.91 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -14.94 |
| Energy contribution | -15.63 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14037871 95 + 27905053 AAGUGAUGCAAGUUUUUACUCAGCAGCAUUGCAGUGGCUGACUUGCCACAGCGUCACACUGAUGAAAAAAACUAUGC----------UACUAUUUACAUUGUAAC .((((..((((((((((..(((.(((...(((.(((((......))))).))).....))).))).))))))).)))----------)))).............. ( -24.90) >DroSec_CAF1 25302 95 + 1 AAGUGAUGCAAGUUUUUACUCAGCAGCAUUGCAGUGGCUGACUUGCCACAGCGUCACACUGAUGAAAAAAACUAUGC----------UACUACUUACUUUCUAUC (((((..((((((((((..(((.(((...(((.(((((......))))).))).....))).))).))))))).)))----------...))))).......... ( -26.50) >DroSim_CAF1 23358 95 + 1 AAGUGAUGCAAGUUUUUACUCAGCAGCAUUGCAGUGGCUGACUUGCCACAGCGUCACACUGAUGAAAAAAACUAUGC----------UACUACUUACUUUCUAUC (((((..((((((((((..(((.(((...(((.(((((......))))).))).....))).))).))))))).)))----------...))))).......... ( -26.50) >DroEre_CAF1 23949 101 + 1 AAGUGAUGCAAGUUUUUACUCUGCAGCACUGCAGUGGCUGACUUGCCACAGC--UACACUGCUGAAAAU-ACUGUUCUGUGAAAGCAUACAAUA-CAAUUCUAUC .((((.(((((((....))).)))).))))((((((((......))).((((--......)))).....-)))))..(((....))).......-.......... ( -27.30) >DroYak_CAF1 29512 102 + 1 AAGUGAUGCAAGUUUUUACUCUGCAGCAUUGCAGUGGCUGUCUUGCCACAGC--UACACUGAUGAAAAU-ACUGUUGUGUGAAACCAUACUAUUUUCAUUCUAGC ..(..((((.((........))...))))..).(((((......))))).((--((....(((((((((-(.....(((((....))))))))))))))).)))) ( -30.00) >DroAna_CAF1 22403 91 + 1 AAGUGAAGCAAGUUUUUCAUCUGCAACCCUGCAGUGGCUUACUUUCCACUUGCUGGCACUGAGAGAAUAAAGGAAGU---------UUAAUAUUUUGUUU----- .((((.(((((((.......(((((....))))).((........)))))))))..))))........(((.(((((---------.....))))).)))----- ( -20.30) >consensus AAGUGAUGCAAGUUUUUACUCAGCAGCAUUGCAGUGGCUGACUUGCCACAGCGUCACACUGAUGAAAAAAACUAUGC__________UACUAUUUACAUUCUAUC ..((((.((.............(((....))).(((((......))))).)).))))................................................ (-14.94 = -15.63 + 0.69)



| Location | 14,037,871 – 14,037,966 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.91 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -14.90 |
| Energy contribution | -17.10 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14037871 95 - 27905053 GUUACAAUGUAAAUAGUA----------GCAUAGUUUUUUUCAUCAGUGUGACGCUGUGGCAAGUCAGCCACUGCAAUGCUGCUGAGUAAAAACUUGCAUCACUU ..................----------(((.(((((((((((.((((((...((.(((((......))))).)).)))))).)))).))))))))))....... ( -29.50) >DroSec_CAF1 25302 95 - 1 GAUAGAAAGUAAGUAGUA----------GCAUAGUUUUUUUCAUCAGUGUGACGCUGUGGCAAGUCAGCCACUGCAAUGCUGCUGAGUAAAAACUUGCAUCACUU ..........((((.((.----------(((.(((((((((((.((((((...((.(((((......))))).)).)))))).)))).)))))))))))).)))) ( -30.50) >DroSim_CAF1 23358 95 - 1 GAUAGAAAGUAAGUAGUA----------GCAUAGUUUUUUUCAUCAGUGUGACGCUGUGGCAAGUCAGCCACUGCAAUGCUGCUGAGUAAAAACUUGCAUCACUU ..........((((.((.----------(((.(((((((((((.((((((...((.(((((......))))).)).)))))).)))).)))))))))))).)))) ( -30.50) >DroEre_CAF1 23949 101 - 1 GAUAGAAUUG-UAUUGUAUGCUUUCACAGAACAGU-AUUUUCAGCAGUGUA--GCUGUGGCAAGUCAGCCACUGCAGUGCUGCAGAGUAAAAACUUGCAUCACUU ....(((..(-((((((.((......))..)))))-)).))).((((..(.--((.(((((......))))).)).)..)))).((((((....)))).)).... ( -30.70) >DroYak_CAF1 29512 102 - 1 GCUAGAAUGAAAAUAGUAUGGUUUCACACAACAGU-AUUUUCAUCAGUGUA--GCUGUGGCAAGACAGCCACUGCAAUGCUGCAGAGUAAAAACUUGCAUCACUU ((.((.(((((((((.(.((........))..).)-))))))))((((((.--((.(((((......))))).)).))))))...........)).))....... ( -26.60) >DroAna_CAF1 22403 91 - 1 -----AAACAAAAUAUUAA---------ACUUCCUUUAUUCUCUCAGUGCCAGCAAGUGGAAAGUAAGCCACUGCAGGGUUGCAGAUGAAAAACUUGCUUCACUU -----..............---------.................((((..(((((((((........)).((((......)))).......))))))).)))). ( -18.30) >consensus GAUAGAAAGUAAAUAGUA__________GCAUAGUUUUUUUCAUCAGUGUGACGCUGUGGCAAGUCAGCCACUGCAAUGCUGCAGAGUAAAAACUUGCAUCACUU ............................(((.((((((((((..(((..(...((.(((((......))))).)).)..)))..))).))))))))))....... (-14.90 = -17.10 + 2.20)

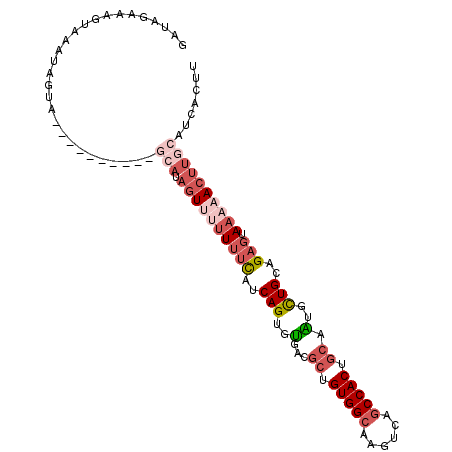

| Location | 14,037,898 – 14,038,005 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 65.23 |
| Mean single sequence MFE | -20.07 |
| Consensus MFE | -8.70 |
| Energy contribution | -9.20 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14037898 107 + 27905053 AUUGCAGUGGCUGACUUGCCACAGCGUCACACUGAUGAAAAAAACUAUGC----------UACUAUUUACAUUGUAACUUAUGAUAUAAUUUGUAUAUCAAAUAAACCUAUCCCAAA- ...((((((((......)))))..((((.....))))..........)))----------.....................(((((((.....))))))).................- ( -19.50) >DroSec_CAF1 25329 107 + 1 AUUGCAGUGGCUGACUUGCCACAGCGUCACACUGAUGAAAAAAACUAUGC----------UACUACUUACUUUCUAUCUUAGGAUAUAAUUUGUAUUUCAAAUAAACCUACCCCAGC- ...((((((((......)))))..((((.....))))..........)))----------...................((((.....(((((.....)))))...)))).......- ( -18.70) >DroSim_CAF1 23385 107 + 1 AUUGCAGUGGCUGACUUGCCACAGCGUCACACUGAUGAAAAAAACUAUGC----------UACUACUUACUUUCUAUCUCAGGAUAUACUUUGUAAUUCAAAUAAACCUACCCCAGC- ...((((((((......)))))..((((.....))))..........)))----------......((((....((((....))))......)))).....................- ( -17.00) >DroEre_CAF1 23976 107 + 1 ACUGCAGUGGCUGACUUGCCACAGC--UACACUGCUGAAAAU-ACUGUUCUGUGAAAGCAUACAAUA-CAAUUCUAUCUUAA-AAAUAC---UUAUAUCAAAAAUUCUUGC--CAAC- ...((((((((......))).((((--......)))).....-)))))..(((....))).......-..............-......---...................--....- ( -17.30) >DroYak_CAF1 29539 108 + 1 AUUGCAGUGGCUGUCUUGCCACAGC--UACACUGAUGAAAAU-ACUGUUGUGUGAAACCAUACUAUUUUCAUUCUAGCUUGA-AAAUAC---CUUUAUCAAAUAAACUUAU--CAUC- ......((((((((......)))))--)))..(((((((...-......(((((....)))))((((((((........)))-))))).---.)))))))...........--....- ( -25.70) >DroAna_CAF1 22430 97 + 1 CCUGCAGUGGCUUACUUUCCACUUGCUGGCACUGAGAGAAUAAAGGAAGU---------UUAAUAUUUUGUUU---------AAUAUCUUUU---UUUCUCCUAAAUUAUCGGUAGUU ((.(((((((........)))).))).)).((((((((((.((((((.((---------(.(((.....))).---------))).))))))---.)))))........))))).... ( -22.20) >consensus AUUGCAGUGGCUGACUUGCCACAGCGUCACACUGAUGAAAAAAACUAUGC__________UACUAUUUACAUUCUAUCUUAAGAUAUACUUUGUAUAUCAAAUAAACCUACCCCAGC_ ...((.(((((......))))).))............................................................................................. ( -8.70 = -9.20 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:42 2006