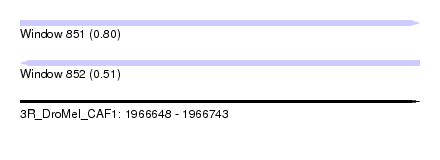
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,966,648 – 1,966,743 |
| Length | 95 |
| Max. P | 0.803830 |

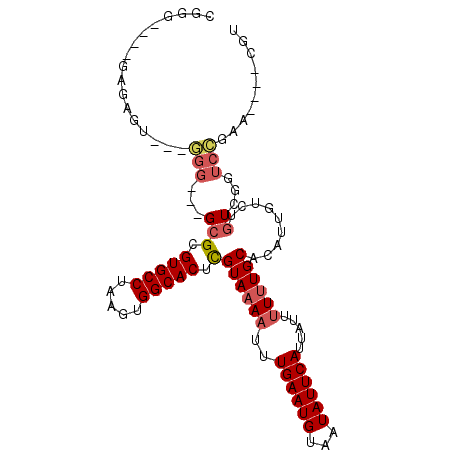
| Location | 1,966,648 – 1,966,743 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.26 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -14.77 |
| Energy contribution | -14.93 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1966648 95 + 27905053 ACA----AUCGGAUGGACAGACAAUGUGCAAAAAAUAAUGAAUAUUGCAUUCAAAUUUUACAAGUGCCACUUAGGCACGCGCACCCCC---ACCCUC---UCCCG ...----...(((.((........(((((.........(((((.....)))))..........(((((.....)))))))))).....---...)).---))).. ( -20.89) >DroVir_CAF1 148468 96 + 1 AGC----UUUUGGCCGACAGACAAUGUGCAAAGAAUAAUGAAUAUUACAUUCAAAUUGUACGAGUGCCACUUAGGCACGCGC---ACACACACACAGAC-UCCC- ...----.((((.....))))...(((((.........(((((.....)))))........(.(((((.....))))).)))---)))...........-....- ( -19.30) >DroPse_CAF1 106621 92 + 1 ACGGGCAGACGGACCGACAGACAAUGUGCAAAAAAUAAUGAAUAUUACAUUCAAAUUUUACGAGUGCCACUUAGGCACGCGC---CCC---GCU-------CCCC ..(((.((.(((..(....).....((((.........(((((.....)))))..........(((((.....)))))))))---.))---)))-------))). ( -24.60) >DroYak_CAF1 129797 95 + 1 ACA----UUCGGACGGACAGACAAUGUGCAAAAAAUAAUGAAUAUUGCAUUCAAAUUUUACGAGUGCCACUUAGGCACGCGC---CCC---ACCCUCUCCCCCCU ...----...((..(((.((.....((((.........(((((.....)))))..........(((((.....)))))))))---...---...)).)))..)). ( -21.20) >DroAna_CAF1 106725 95 + 1 ACGG-CGAACGUACAGACAGACAAUGUGCAAAAAAUAAUGAAUAUUACAUUCAAAUUUUACGAGUGCCACUUAGGCACGCCC---CCG---CAUUUC---GCCCG ..((-((((.(((((.........))))).........(((((.....)))))..........(((((.....)))))((..---..)---)..)))---))).. ( -24.10) >DroPer_CAF1 107750 92 + 1 ACGGACAGACGGACCGACAGACAAUGUGCAAAAAAUAAUGAAUAUUACAUUCAAAUUUUACGAGUGCCACUUAGGCACGCGC---CCC---GCU-------CCCC ..((..((.(((..(....).....((((.........(((((.....)))))..........(((((.....)))))))))---.))---)))-------..)) ( -20.10) >consensus ACG____AACGGACCGACAGACAAUGUGCAAAAAAUAAUGAAUAUUACAUUCAAAUUUUACGAGUGCCACUUAGGCACGCGC___CCC___ACUCUC____CCCC .........................((((.........(((((.....)))))..........(((((.....)))))))))....................... (-14.77 = -14.93 + 0.17)

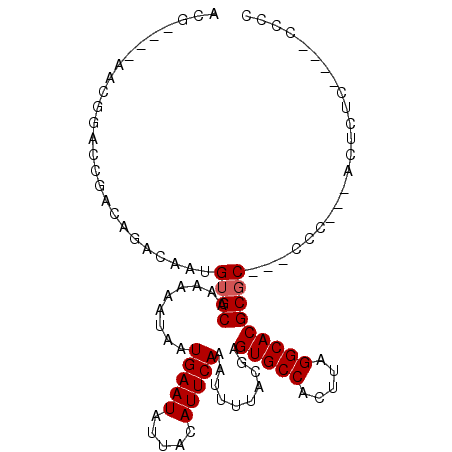
| Location | 1,966,648 – 1,966,743 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.26 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -14.84 |
| Energy contribution | -15.48 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1966648 95 - 27905053 CGGGA---GAGGGU---GGGGGUGCGCGUGCCUAAGUGGCACUUGUAAAAUUUGAAUGCAAUAUUCAUUAUUUUUUGCACAUUGUCUGUCCAUCCGAU----UGU ...((---..((((---(((.(.((..(((((.....))))).(((((((..((((((...)))))).....)))))))....)).).)))))))..)----).. ( -28.50) >DroVir_CAF1 148468 96 - 1 -GGGA-GUCUGUGUGUGUGU---GCGCGUGCCUAAGUGGCACUCGUACAAUUUGAAUGUAAUAUUCAUUAUUCUUUGCACAUUGUCUGUCGGCCAAAA----GCU -((.(-(.(.((((((.(((---(((.(((((.....))))).))))))...((((((...)))))).........)))))).).)).))(((.....----))) ( -31.80) >DroPse_CAF1 106621 92 - 1 GGGG-------AGC---GGG---GCGCGUGCCUAAGUGGCACUCGUAAAAUUUGAAUGUAAUAUUCAUUAUUUUUUGCACAUUGUCUGUCGGUCCGUCUGCCCGU (((.-------(((---(((---.((((((((.....)))))..((((((..((((((...)))))).....)))))).........).)).)))).)).))).. ( -33.00) >DroYak_CAF1 129797 95 - 1 AGGGGGGAGAGGGU---GGG---GCGCGUGCCUAAGUGGCACUCGUAAAAUUUGAAUGCAAUAUUCAUUAUUUUUUGCACAUUGUCUGUCCGUCCGAA----UGU .(((.(((.(((((---(..---(((.(((((.....))))).)))..........(((((.............))))))))..))).))).)))...----... ( -30.32) >DroAna_CAF1 106725 95 - 1 CGGGC---GAAAUG---CGG---GGGCGUGCCUAAGUGGCACUCGUAAAAUUUGAAUGUAAUAUUCAUUAUUUUUUGCACAUUGUCUGUCUGUACGUUCG-CCGU ..(((---(((.((---(((---(((((((((.....)))))..((((((..((((((...)))))).....)))))).....))))..)))))..))))-)).. ( -32.60) >DroPer_CAF1 107750 92 - 1 GGGG-------AGC---GGG---GCGCGUGCCUAAGUGGCACUCGUAAAAUUUGAAUGUAAUAUUCAUUAUUUUUUGCACAUUGUCUGUCGGUCCGUCUGUCCGU (((.-------(((---(((---.((((((((.....)))))..((((((..((((((...)))))).....)))))).........).)).)))).)).))).. ( -30.60) >consensus CGGG____GAGAGU___GGG___GCGCGUGCCUAAGUGGCACUCGUAAAAUUUGAAUGUAAUAUUCAUUAUUUUUUGCACAUUGUCUGUCGGUCCGAA____CGU .................(((...(((.(((((.....))))).)((((((..((((((...)))))).....)))))).........))...))).......... (-14.84 = -15.48 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:26 2006