
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,023,549 – 14,023,668 |
| Length | 119 |
| Max. P | 0.999933 |

| Location | 14,023,549 – 14,023,647 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -29.74 |
| Energy contribution | -30.66 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14023549 98 + 27905053 UCCCUUCUCCCUCGUUCCACUCAGGUGAGUGAGGAGCAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUACACAUUGUACACAGGUAGGUUGCUACCU ...((((((.((((..((.....)))))).))))))...((((((.((..((.((((....)))).))..))))))))....((((((....)))))) ( -32.30) >DroSec_CAF1 8647 98 + 1 UCCCUCCUCCCUCGUUCCACUCAGGUGAGUGGGGAACAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUACACAUUGUACACAGGUAGGUUGCUACCU ((((..((((((.(.......)))).)))..))))....((((((.((..((.((((....)))).))..))))))))....((((((....)))))) ( -35.10) >DroSim_CAF1 8553 98 + 1 UCCCUCCUCCCUCGUUCCACUCAGGUGAGUGAGGAACAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUACACAUUGUACACAGGUAGGUUGCUACCU ..((((((((((.(.......)))).))).)))).....((((((.((..((.((((....)))).))..))))))))....((((((....)))))) ( -32.90) >DroEre_CAF1 8628 98 + 1 UCCCUCCUACCUCGUUCCGCACAGGUGAGUGAGGAGUAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUACACAUUGUACACAGGUAGGUUUCUACCU ..((((((((((.((.....)))))).)).)))).((..((((((.((..((.((((....)))).))..))))))))..))((((((....)))))) ( -33.40) >DroYak_CAF1 8581 97 + 1 UUCCCCCUACCUCGUUCCGCACAGGUGAGUGAG-UGUAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUUCACAUUGUACACAGGUAGGUUGCUACCU ......(((((((((((.((....)))))))))-.))))((((((.((..((.((((....)))).))..))))))))....((((((....)))))) ( -30.80) >consensus UCCCUCCUCCCUCGUUCCACUCAGGUGAGUGAGGAGCAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUACACAUUGUACACAGGUAGGUUGCUACCU ...((((((.((((..((.....)))))).))))))...((((((.((..((.((((....)))).))..))))))))....((((((....)))))) (-29.74 = -30.66 + 0.92)


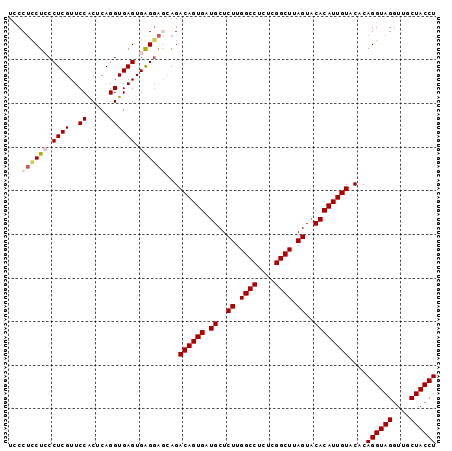
| Location | 14,023,549 – 14,023,647 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -25.64 |
| Energy contribution | -26.80 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14023549 98 - 27905053 AGGUAGCAACCUACCUGUGUACAAUGUGUACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUGCUCCUCACUCACCUGAGUGGAACGAGGGAGAAGGGA ((((((....))))))....(((.(((((.((..(((....)))..)).))).)).)))((.((((((((((....)))))......))))).))... ( -30.60) >DroSec_CAF1 8647 98 - 1 AGGUAGCAACCUACCUGUGUACAAUGUGUACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUGUUCCCCACUCACCUGAGUGGAACGAGGGAGGAGGGA ((((((....))))))....(((.(((((.((..(((....)))..)).))).)).)))((.((((((((((....)))))......))))).))... ( -32.30) >DroSim_CAF1 8553 98 - 1 AGGUAGCAACCUACCUGUGUACAAUGUGUACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUGUUCCUCACUCACCUGAGUGGAACGAGGGAGGAGGGA ((((((....))))))....(((.(((((.((..(((....)))..)).))).)).)))...((((((.(((.(((..(.....).)))))))))))) ( -32.10) >DroEre_CAF1 8628 98 - 1 AGGUAGAAACCUACCUGUGUACAAUGUGUACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUACUCCUCACUCACCUGUGCGGAACGAGGUAGGAGGGA ((((((....))))))(((.(((.(((((.((..(((....)))..)).))).)).))).)))(((((.((.((((((.....)).))))))))))). ( -34.60) >DroYak_CAF1 8581 97 - 1 AGGUAGCAACCUACCUGUGUACAAUGUGAACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUACA-CUCACUCACCUGUGCGGAACGAGGUAGGGGGAA ......(..((((((((((((....((((.((..(((....)))....))..))))....))))-).(((......))).......)))))))..).. ( -29.90) >consensus AGGUAGCAACCUACCUGUGUACAAUGUGUACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUGCUCCUCACUCACCUGAGUGGAACGAGGGAGGAGGGA ((((((....))))))....(((.(((((.((..(((....)))..)).))).)).)))...((((((.(((.((.....))...))).))))))... (-25.64 = -26.80 + 1.16)



| Location | 14,023,571 – 14,023,668 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -29.84 |
| Energy contribution | -30.84 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14023571 97 + 27905053 AGGUGAGUGAGGAGCAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUACACAUUGUACACAGGUAGGUUGCUACCUAAAAGCCUAAAGGCCAGCGGG ......((.(((((((........)))))))(((((...(((((.((((.....))))..((((((....))))))..)))))...))))).))... ( -36.90) >DroSec_CAF1 8669 97 + 1 AGGUGAGUGGGGAACAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUACACAUUGUACACAGGUAGGUUGCUACCUAAAAGCCUAAAGGCCAGCAGG ....(((((....((.....))..)))))(((((((...(((((.((((.....))))..((((((....))))))..)))))...))))))).... ( -32.70) >DroSim_CAF1 8575 97 + 1 AGGUGAGUGAGGAACAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUACACAUUGUACACAGGUAGGUUGCUACCUAAAAGCCUAAAGGCCAGCGGG .(((..(.((((...(((((....)).)))...)))).)(((((.((((.....))))..((((((....))))))..))))).....)))...... ( -32.30) >DroEre_CAF1 8650 97 + 1 AGGUGAGUGAGGAGUAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUACACAUUGUACACAGGUAGGUUUCUACCUAAAUGCCUAAAGGCCAGCGGG ......((.(((((((........)))))))(((((...(((...((((.....))))..((((((....))))))....)))...))))).))... ( -32.40) >DroYak_CAF1 8603 96 + 1 AGGUGAGUGAG-UGUAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUUCACAUUGUACACAGGUAGGUUGCUACCUAACAGCCUUAAGGCCAGCGGG .(.((.((..(-(((..((((((.((..((.((((....)))).))..))))))))))))((((((....)))))).)).(((....))))).)... ( -31.10) >consensus AGGUGAGUGAGGAGCAGACAGUGAUGCUCUUGGCCUCUCGGCUUAGUACACAUUGUACACAGGUAGGUUGCUACCUAAAAGCCUAAAGGCCAGCGGG ......((.(((((((........)))))))(((((...(((((.((((.....))))..((((((....))))))..)))))...))))).))... (-29.84 = -30.84 + 1.00)


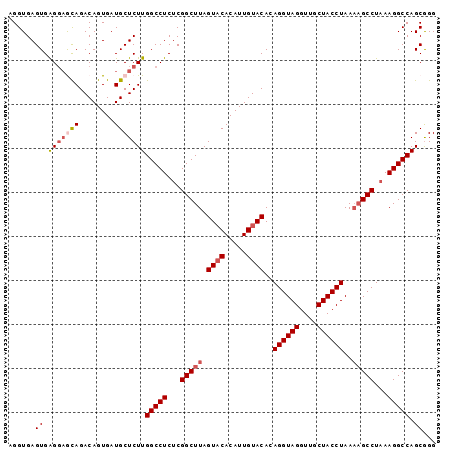
| Location | 14,023,571 – 14,023,668 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -29.74 |
| Energy contribution | -30.54 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.65 |
| SVM RNA-class probability | 0.999933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14023571 97 - 27905053 CCCGCUGGCCUUUAGGCUUUUAGGUAGCAACCUACCUGUGUACAAUGUGUACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUGCUCCUCACUCACCU .....((((((((.(((((.(((((((....))))))).((((.....)))).))))).)))))))).(((((........)))))........... ( -36.50) >DroSec_CAF1 8669 97 - 1 CCUGCUGGCCUUUAGGCUUUUAGGUAGCAACCUACCUGUGUACAAUGUGUACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUGUUCCCCACUCACCU .....((((((((.(((((.(((((((....))))))).((((.....)))).))))).)))))))).(((((........)))))........... ( -34.10) >DroSim_CAF1 8575 97 - 1 CCCGCUGGCCUUUAGGCUUUUAGGUAGCAACCUACCUGUGUACAAUGUGUACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUGUUCCUCACUCACCU .....((((((((.(((((.(((((((....))))))).((((.....)))).))))).)))))))).(((((........)))))........... ( -34.10) >DroEre_CAF1 8650 97 - 1 CCCGCUGGCCUUUAGGCAUUUAGGUAGAAACCUACCUGUGUACAAUGUGUACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUACUCCUCACUCACCU ...((((((((((.(((...(((((((....))))))).((((.....))))...))).)))))))...)))......................... ( -32.00) >DroYak_CAF1 8603 96 - 1 CCCGCUGGCCUUAAGGCUGUUAGGUAGCAACCUACCUGUGUACAAUGUGAACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUACA-CUCACUCACCU ...(((((((((..((((..(((((((....)))))))........(....)..))))..))))))...))).............-........... ( -27.30) >consensus CCCGCUGGCCUUUAGGCUUUUAGGUAGCAACCUACCUGUGUACAAUGUGUACUAAGCCGAGAGGCCAAGAGCAUCACUGUCUGCUCCUCACUCACCU ...((((((((((.(((((.(((((((....))))))).((((.....)))).))))).)))))))...)))......................... (-29.74 = -30.54 + 0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:36 2006