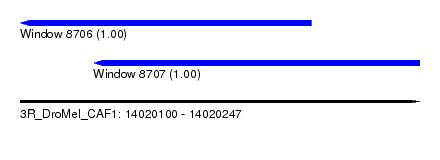
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,020,100 – 14,020,247 |
| Length | 147 |
| Max. P | 0.999920 |

| Location | 14,020,100 – 14,020,207 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.08 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -27.32 |
| Energy contribution | -27.45 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14020100 107 - 27905053 GCGGUCGAAUAUUCGUAAGUCGGACUAGAUCUAUGUAACGAAUAUAGUUCGCCUUGCGGACAUUCGACUGUUUGCUUAAAGUAAUGUUAAACAUGUAUGCAACAAGU ((((((((((.((((((((.(((((((..((........))...))))))).)))))))).))))))))))((((.((.....(((.....))).)).))))..... ( -36.10) >DroSec_CAF1 5249 93 - 1 GCGGUCGAACAUCAGUAAGUCGAACUAGAUCUA-----CGAAUAUAGUUCGCCUUGCGGAGGUUCGUCUGUGUGCUUAAAGUAAU---------GUAUGCAACAAGU ((((.(((((.((.(((((.(((((((......-----......))))))).))))).)).))))).)))).(((....(....)---------....)))...... ( -29.30) >DroSim_CAF1 5228 102 - 1 GCGGUCGAGCAUCAGUAAGUCGAACUAGAUCUA-----CGAAUAUAGUUCGCCUUGCGGAGGUUCGUCUGUGUGCUUAAAGUAAUGUUAAACAUGUAUGCAACAAGU ((((.(((((.((.(((((.(((((((......-----......))))))).))))).)).))))).)))).(((.((.....(((.....))).)).)))...... ( -30.00) >DroEre_CAF1 5241 100 - 1 GCGGUCGAGCUUCCGUAAGUCGGACCACAACUA-----G-AA-ACAGUUCGCGGUAUGGACGUUCGACUGUAUGCUUAAAGUAAUGUUAAACAUGUAUGCAACAAGU ((((((((((.((((((.(.(((((........-----.-..-...))))))..)))))).)))))))))).(((.((.....(((.....))).)).)))...... ( -29.94) >DroYak_CAF1 5209 100 - 1 GCGGCCGAGCUUCUGUAAGUCGGACUAGAACUA-----G-AA-UUAGUUCGUCUUAUGGACGUUCGACUGUAUGCUUAAAGUAAUGUUAAACAUGUAUGCAACAAGA ((((.(((((.((((((((.((((((((.....-----.-..-)))))))).)))))))).))))).)))).(((.((.....(((.....))).)).)))...... ( -31.40) >consensus GCGGUCGAGCAUCAGUAAGUCGGACUAGAUCUA_____CGAAUAUAGUUCGCCUUGCGGACGUUCGACUGUAUGCUUAAAGUAAUGUUAAACAUGUAUGCAACAAGU ((((.(((((.((((((((.(((((((.................))))))).)))))))).))))).)))).(((.((.....(((.....))).)).)))...... (-27.32 = -27.45 + 0.13)



| Location | 14,020,127 – 14,020,247 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.82 |
| Mean single sequence MFE | -33.79 |
| Consensus MFE | -31.05 |
| Energy contribution | -30.77 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14020127 120 - 27905053 ACCAAGCAACAAAAACUGUAAUCGUAAUGAACAAACAAAUGCGGUCGAAUAUUCGUAAGUCGGACUAGAUCUAUGUAACGAAUAUAGUUCGCCUUGCGGACAUUCGACUGUUUGCUUAAA ...((((((..............((.....))........((((((((((.((((((((.(((((((..((........))...))))))).)))))))).))))))))))))))))... ( -37.80) >DroSec_CAF1 5267 115 - 1 ACCAAGCAACAAAAACAGUAAUCGUAAUGAACAAACAAAUGCGGUCGAACAUCAGUAAGUCGAACUAGAUCUA-----CGAAUAUAGUUCGCCUUGCGGAGGUUCGUCUGUGUGCUUAAA ...((((................((.....))......((((((.(((((.((.(((((.(((((((......-----......))))))).))))).)).))))).))))))))))... ( -32.00) >DroSim_CAF1 5255 115 - 1 ACCAAGCAACAAAAACAGUAAUCGUAAUGAACAAACAAAUGCGGUCGAGCAUCAGUAAGUCGAACUAGAUCUA-----CGAAUAUAGUUCGCCUUGCGGAGGUUCGUCUGUGUGCUUAAA ...((((................((.....))......((((((.(((((.((.(((((.(((((((......-----......))))))).))))).)).))))).))))))))))... ( -32.00) >DroEre_CAF1 5268 113 - 1 ACCAAGCAACAAAAACAGUAAACGUAAUGAACAAACAAAUGCGGUCGAGCUUCCGUAAGUCGGACCACAACUA-----G-AA-ACAGUUCGCGGUAUGGACGUUCGACUGUAUGCUUAAA ...((((................((.....))......((((((((((((.((((((.(.(((((........-----.-..-...))))))..)))))).))))))))))))))))... ( -32.84) >DroYak_CAF1 5236 113 - 1 ACCAAGCAACAAAAACAGUAAUCGUAAUGAACAAACAAAUGCGGCCGAGCUUCUGUAAGUCGGACUAGAACUA-----G-AA-UUAGUUCGUCUUAUGGACGUUCGACUGUAUGCUUAAA ...((((................((.....))......((((((.(((((.((((((((.((((((((.....-----.-..-)))))))).)))))))).))))).))))))))))... ( -34.30) >consensus ACCAAGCAACAAAAACAGUAAUCGUAAUGAACAAACAAAUGCGGUCGAGCAUCAGUAAGUCGGACUAGAUCUA_____CGAAUAUAGUUCGCCUUGCGGACGUUCGACUGUAUGCUUAAA ...((((................((.....))......((((((.(((((.((((((((.(((((((.................))))))).)))))))).))))).))))))))))... (-31.05 = -30.77 + -0.28)

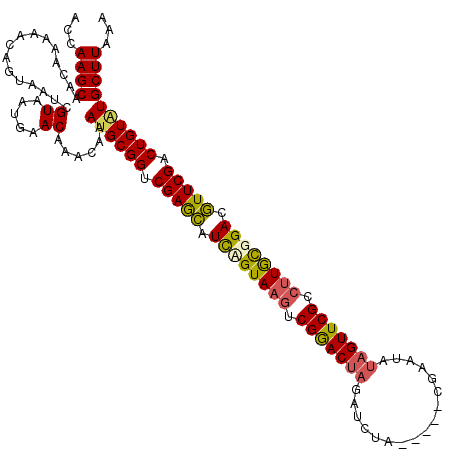

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:24 2006