| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,014,976 – 14,015,093 |
| Length | 117 |
| Max. P | 0.864513 |

| Location | 14,014,976 – 14,015,093 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.94 |
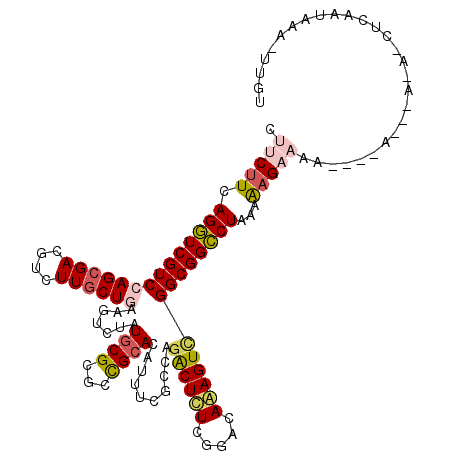
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.73 |
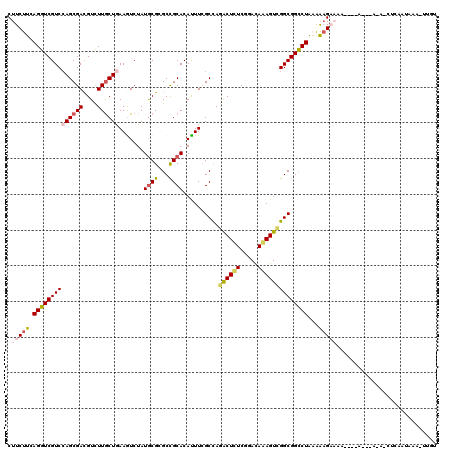
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
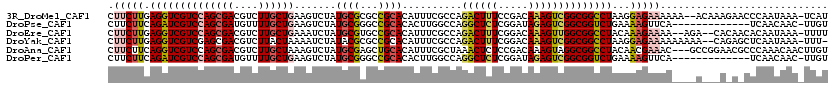
>3R_DroMel_CAF1 14014976 117 - 27905053 CUUCUUGAGGUCGUCCAGCGACGUCUUGCUGAAGUCUAUGCGCGCCGCACAUUUCGCCAGACUUUCCGACAAAGUCGGCGGCCUAAGGAGAAAAAA--ACAAAGAACCCAAUAAA-UCAU ((((((.((((((((....)))((((.((.(((((...((((...)))).))))))).))))...(((((...))))).)))))))))))......--.................-.... ( -32.40) >DroPse_CAF1 130 106 - 1 CUUCUUCAGAUCGUCCAGCGAUGUUUUGCUGAAGUCUAUGCGGGCCGCACACUUGGCCAGGCUCUCGGAUAGAGUCGGCGGUCUGAAAAGUUCA-------------UCAACAAC-UUGU ....(((((((((((((((((....))))))...........(((((......))))).((((((.....)))))))))))))))))(((((..-------------.....)))-)).. ( -38.50) >DroEre_CAF1 133 115 - 1 CUUCUUGAGGUCGUCCAGCGACGUCUUGCUGAAAUCUAUGCGUGCCGCACAUUUCGCCAGACUUUCGGACAAAGUUGGCGGCCUACAAAGAAAA--AGA--CACAACACAAUAAA-UUUU .(((((.((((((.(((((...((((.((.(((((...((((...)))).))))))).))))(((.....))))))))))))))...)))))..--...--..............-.... ( -31.20) >DroYak_CAF1 132 116 - 1 CUUCUUGAGGUCGUCGAGCGACGUCUUACUAAAAUCUAUACGCGCCGCACAUUUCGCCAGACUUUCGGACAAAGUCGGCGGCCUAAGGAGAAAAAAAAA--CAGAGCUCAAUAAA-UUU- ((((((.(((((((((.(((.(((..((........)).)))))))).)).....(((.((((((.....)))))))))))))))))))).........--..............-...- ( -31.40) >DroAna_CAF1 126 117 - 1 CUUCUUCAGGUCGUCCAGCGACGUCUUGCUAAAGUCUAUGCGAGCUGCACAUUUCGCUAAACUCUCCGACAAAGUAGGCGGCCUACAACGAAAC---GCCGGAACGCCCAAACAACUUGU ........((.(((((.(((..(((.......(((.((.(((((........))))))).)))....)))...(((((...))))).......)---)).)).))).))........... ( -26.20) >DroPer_CAF1 130 106 - 1 CUUCUUCAGAUCGUCCAGCGAUGUUUUGCUGAAGUCUAUGCGGGCCGCACACUUGGCCAGGCUCUCGGAUAGAGUCGGCGGUCUGAAAAGUUCA-------------UCAACAAC-UUGU ....(((((((((((((((((....))))))...........(((((......))))).((((((.....)))))))))))))))))(((((..-------------.....)))-)).. ( -38.50) >consensus CUUCUUCAGGUCGUCCAGCGACGUCUUGCUGAAGUCUAUGCGCGCCGCACAUUUCGCCAGACUCUCGGACAAAGUCGGCGGCCUAAAAAGAAAA____A___A_A_CUCAAUAAA_UUGU .(((((.((((((((((((((....)))))).......((((...))))..........((((((.....))))))))))))))...)))))............................ (-22.37 = -22.73 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:21 2006