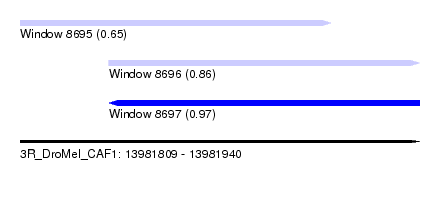
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,981,809 – 13,981,940 |
| Length | 131 |
| Max. P | 0.967107 |

| Location | 13,981,809 – 13,981,911 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.03 |
| Mean single sequence MFE | -20.87 |
| Consensus MFE | -12.24 |
| Energy contribution | -12.14 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13981809 102 + 27905053 CAUAUAGUGUAAACAUAAUAAUCAACGGGCAUUAUAUGCCGAUAUUUGCCGAUAUAUAUAUAUUACAUACAUAUGCAUGUGGCGAUCAGCAGAAUGCGCUUU ..(((.(((....))).)))......(.(((((...(((.((...(((((.((((.(((((.........))))).))))))))))).))).))))).)... ( -21.10) >DroEre_CAF1 243506 101 + 1 CAUAUAGUGUAAACAUAAUAAUCAACGGGCAUUAUAUGCCUACAUAUACCUAUGUAUAUACAU-ACGAGUAUAUGUAUGUGGCGAUCAGCAGAAUGCGCUUU .....((((((..((((.........((((((...))))))((((((((((((((....))))-).).)))))))).))))((.....))....)))))).. ( -26.70) >DroYak_CAF1 264907 81 + 1 CAUAUAGUGUAAACAUAAUAAUCAACGUACAUUAUAUGCCGA--UAUACCU-------------AC------AACUAUGUGGCGAUCAGCAGAAUGCGCUUU ..(((.(((....))).)))......(..((((...(((.((--(....((-------------((------(....)))))..))).))).))))..)... ( -14.80) >consensus CAUAUAGUGUAAACAUAAUAAUCAACGGGCAUUAUAUGCCGA_AUAUACCUAU_UAUAUA_AU_AC____AUAUGUAUGUGGCGAUCAGCAGAAUGCGCUUU ((((((((((..................))))))))))....................................((((...((.....))...))))..... (-12.24 = -12.14 + -0.11)


| Location | 13,981,838 – 13,981,940 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -21.28 |
| Energy contribution | -23.23 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13981838 102 + 27905053 CAUUAUAUGCCGAUAUUUGCCGAUAUAUAUAUAUUACAUACAUAUGCAUGUGGCGAUCAGCAGAAUGCGCUUUUGCAAAAUGGCCGGCGGUCAGCGCGGCCC ((((...(((.((...(((((.((((.(((((.........))))).))))))))))).))).)))).((....)).....(((((((.....)).))))). ( -28.30) >DroEre_CAF1 243535 101 + 1 CAUUAUAUGCCUACAUAUACCUAUGUAUAUACAU-ACGAGUAUAUGUAUGUGGCGAUCAGCAGAAUGCGCUUUUGCAAAAUGGCCGGCGGUCAGCGCGGCCC .......(((((((((((((((((((....))))-).).)))))))))...))))....((((((.....)))))).....(((((((.....)).))))). ( -34.60) >DroYak_CAF1 264936 81 + 1 CAUUAUAUGCCGA--UAUACCU-------------AC------AACUAUGUGGCGAUCAGCAGAAUGCGCUUUUGCAAAAUGGCCGGCGGUCAGCGCGGCCC ((((...(((.((--(....((-------------((------(....)))))..))).))).)))).((....)).....(((((((.....)).))))). ( -25.20) >consensus CAUUAUAUGCCGA_AUAUACCUAU_UAUAUA_AU_AC____AUAUGUAUGUGGCGAUCAGCAGAAUGCGCUUUUGCAAAAUGGCCGGCGGUCAGCGCGGCCC .......((((...((((((....((((((((.....))))))))))))))))))....((((((.....)))))).....(((((((.....)).))))). (-21.28 = -23.23 + 1.95)



| Location | 13,981,838 – 13,981,940 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -21.79 |
| Energy contribution | -22.80 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13981838 102 - 27905053 GGGCCGCGCUGACCGCCGGCCAUUUUGCAAAAGCGCAUUCUGCUGAUCGCCACAUGCAUAUGUAUGUAAUAUAUAUAUAUCGGCAAAUAUCGGCAUAUAAUG .(((((.((.....)))))))....(((......)))...(((((((.(((((((((....)))))).((((....)))).)))....)))))))....... ( -31.40) >DroEre_CAF1 243535 101 - 1 GGGCCGCGCUGACCGCCGGCCAUUUUGCAAAAGCGCAUUCUGCUGAUCGCCACAUACAUAUACUCGU-AUGUAUAUACAUAGGUAUAUGUAGGCAUAUAAUG .(((((.((.....))))))).....((....))((.....)).....(((...((((((((((..(-((((....))))))))))))))))))........ ( -32.40) >DroYak_CAF1 264936 81 - 1 GGGCCGCGCUGACCGCCGGCCAUUUUGCAAAAGCGCAUUCUGCUGAUCGCCACAUAGUU------GU-------------AGGUAUA--UCGGCAUAUAAUG .(((((.((.....)))))))....(((......)))...(((((((.((((((....)------))-------------.)))..)--))))))....... ( -27.60) >consensus GGGCCGCGCUGACCGCCGGCCAUUUUGCAAAAGCGCAUUCUGCUGAUCGCCACAUACAUAU____GU_AU_UAUAUA_AUAGGUAUAU_UCGGCAUAUAAUG .(((((.((.....)))))))....(((......)))...(((((((.(((......((((((.......)))))).....)))....)))))))....... (-21.79 = -22.80 + 1.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:14 2006