
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,969,808 – 13,969,911 |
| Length | 103 |
| Max. P | 0.571271 |

| Location | 13,969,808 – 13,969,911 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
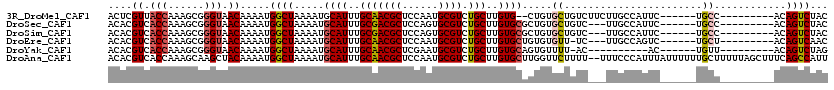
>3R_DroMel_CAF1 13969808 103 + 27905053 ACUCGUUACCAAAGCGGGUAACAAAAUGGCUAAAAUGCAUUUGCAACGCUCCAAUGCGUCUGCUUGUG--CUGUGCUGUCUUCUUGCCAUUC------UGCC---------ACAGUCUAC ....((((((......))))))....((((......((((..(((((((......)))).)))..)))--)((.((.........))))...------.)))---------)........ ( -29.40) >DroSec_CAF1 240346 102 + 1 ACACGUCACCAAAGCGGGUAACAAAAUGGCUAAAAUGCAUUUGCGACGCUCCAGUGCGUCUGCUUGUGCGCUGUGCUGUC---UUGCCAUUC------UGCC---------ACAGUCUAC ...(((.......)))((((....((((((......((((..(((((((......))))).((....)))).))))....---..)))))).------))))---------......... ( -30.00) >DroSim_CAF1 249658 102 + 1 ACACGUCACCAAAGCGGGUAACAAAAUGGCUAAAAUGCAUUUGCGACGCUCCAGUGCGUCUGCUUGUGCGCUGUGCUGUC---UUGCCAUUC------UGCC---------ACAGUCUAC ...(((.......)))((((....((((((......((((..(((((((......))))).((....)))).))))....---..)))))).------))))---------......... ( -30.00) >DroEre_CAF1 231841 101 + 1 ACACGUCACCAAAGCGGGUAACAAAAUGGCUAAAAUGCAUUUGCAACGCUCCAAUGCGUCUGCUUGUGCUGUGUGUU-UC---UUGCCAGUC------UGCU---------ACAGUCAAC ....((.(((......))).))....((((..((((((((..(((((((......)))).)))..)))).....)))-).---..))))(.(------((..---------.))).)... ( -24.70) >DroYak_CAF1 253341 94 + 1 ACACGUCACCAAAGCGGGUAACAAAAUGGCUAAAAUGCAUUUGCAACGCUCGAAUGCGUCUGCUUGUGCAGUGUUUU-AC----------AC------UGUU---------ACAGUCUAG ....((.(((......))).)).....((((.....((((..(((((((......)))).)))..))))((((....-.)----------))------)...---------..))))... ( -25.90) >DroAna_CAF1 233202 118 + 1 ACACGUCACCAAAGCAAGCUACAAAAUGGCUAAAAUGCAUUUGCAACGCUCCAAUGCGUCUGCUUGUGCUUGGUUCUUUU--UUUCCCAUUUAUUUUUUGCUUUUUAGCUUUCAGCCAUU ..........((((((((.....((((((..((((.((((..(((((((......)))).)))..)))).........))--))..))))))....))))))))...((.....)).... ( -26.00) >consensus ACACGUCACCAAAGCGGGUAACAAAAUGGCUAAAAUGCAUUUGCAACGCUCCAAUGCGUCUGCUUGUGCGCUGUGCUGUC___UUGCCAUUC______UGCC_________ACAGUCUAC ....((.(((......))).)).....((((.....((((..(((((((......)))).)))..)))).....((.......................))............))))... (-17.47 = -17.72 + 0.25)
| Location | 13,969,808 – 13,969,911 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -19.29 |
| Energy contribution | -19.18 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
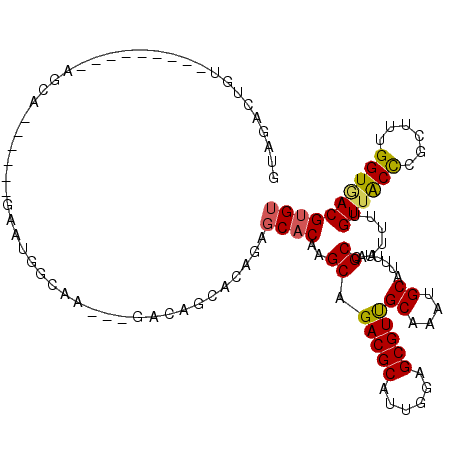
>3R_DroMel_CAF1 13969808 103 - 27905053 GUAGACUGU---------GGCA------GAAUGGCAAGAAGACAGCACAG--CACAAGCAGACGCAUUGGAGCGUUGCAAAUGCAUUUUAGCCAUUUUGUUACCCGCUUUGGUAACGAGU .......((---------((((------(((((((.((((..((((...)--)....((((.(((......)))))))...))..)))).)))))))))))))....((((....)))). ( -29.50) >DroSec_CAF1 240346 102 - 1 GUAGACUGU---------GGCA------GAAUGGCAA---GACAGCACAGCGCACAAGCAGACGCACUGGAGCGUCGCAAAUGCAUUUUAGCCAUUUUGUUACCCGCUUUGGUGACGUGU ..((...((---------((((------(((((((.(---((..(((..((((....)).(((((......)))))))...)))..))).)))))))))))))...))..(....).... ( -35.00) >DroSim_CAF1 249658 102 - 1 GUAGACUGU---------GGCA------GAAUGGCAA---GACAGCACAGCGCACAAGCAGACGCACUGGAGCGUCGCAAAUGCAUUUUAGCCAUUUUGUUACCCGCUUUGGUGACGUGU ..((...((---------((((------(((((((.(---((..(((..((((....)).(((((......)))))))...)))..))).)))))))))))))...))..(....).... ( -35.00) >DroEre_CAF1 231841 101 - 1 GUUGACUGU---------AGCA------GACUGGCAA---GA-AACACACAGCACAAGCAGACGCAUUGGAGCGUUGCAAAUGCAUUUUAGCCAUUUUGUUACCCGCUUUGGUGACGUGU .......((---------((((------((.((((..---(.-....)...(((...((((.(((......)))))))...)))......)))).)))))))).(((....)))...... ( -26.10) >DroYak_CAF1 253341 94 - 1 CUAGACUGU---------AACA------GU----------GU-AAAACACUGCACAAGCAGACGCAUUCGAGCGUUGCAAAUGCAUUUUAGCCAUUUUGUUACCCGCUUUGGUGACGUGU (((((.(((---------(.((------((----------(.-....))))).....((((.(((......)))))))...)))).))))).......((((((......)))))).... ( -27.00) >DroAna_CAF1 233202 118 - 1 AAUGGCUGAAAGCUAAAAAGCAAAAAAUAAAUGGGAAA--AAAAGAACCAAGCACAAGCAGACGCAUUGGAGCGUUGCAAAUGCAUUUUAGCCAUUUUGUAGCUUGCUUUGGUGACGUGU ..(((((....(((....)))..........(((....--.......)))))).)).....((((((..((((((((((((.((......))...))))))))..))))..))).))).. ( -30.20) >consensus GUAGACUGU_________AGCA______GAAUGGCAA___GACAGCACAGAGCACAAGCAGACGCAUUGGAGCGUUGCAAAUGCAUUUUAGCCAUUUUGUUACCCGCUUUGGUGACGUGU ...................................................((((..((.(((((......)))))((....))......))......((((((......)))))))))) (-19.29 = -19.18 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:07 2006