
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,854,594 – 13,854,742 |
| Length | 148 |
| Max. P | 0.761144 |

| Location | 13,854,594 – 13,854,684 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 86.59 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -20.19 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13854594 90 + 27905053 AGAGCUCUAAGUGCAUGGAAUUUAUCAUUGACGAUGGUCGACGAUCGUCGUCAGUGGCGGUAA-----AGGAAGUCGGCGGUGGAGGCGUUUGCC ...((((((......))))......((((((((((((((...))))))))))))))))(((((-----(.(...(((....)))...).)))))) ( -29.10) >DroSec_CAF1 125777 84 + 1 AGAGCUCUAAGUGCAUGGAAUUUAUCAUUGACGAUGG------AUCGUCGUCAGUGGCGGUGU-----GUGGAGUCGAAGGUGGAGGCGUUUGCC .((.(((((...((((.(......((((((((((((.------..))))))))))))).))))-----.)))))))...((..((....))..)) ( -26.90) >DroSim_CAF1 131801 84 + 1 AGAGCUCUAAGUGCAUGGAAUUUAUCAUUGACGAUGG------AUCGUCGUCAGUGGCGGUGU-----GUGGAGUCGAAGGUGGAGGCGUUUGCC .((.(((((...((((.(......((((((((((((.------..))))))))))))).))))-----.)))))))...((..((....))..)) ( -26.90) >DroYak_CAF1 136784 89 + 1 AGAGCUCUAAGUGCAUGGAAUUUAUCAUUGCCGUUGG------AUCGUCGUCAGUGGCGAUGGCGGUGGUGGAGUUAGAGGUGGAGGCGUUUGCC ..((((((((((.......))))).(((..(((((..------(((((((....))))))))))))..))))))))...((..((....))..)) ( -27.80) >consensus AGAGCUCUAAGUGCAUGGAAUUUAUCAUUGACGAUGG______AUCGUCGUCAGUGGCGGUGU_____GUGGAGUCGAAGGUGGAGGCGUUUGCC ...((((((............(..(((((((((((((.......)))))))))))))..).........))))))....((..((....))..)) (-20.19 = -21.00 + 0.81)

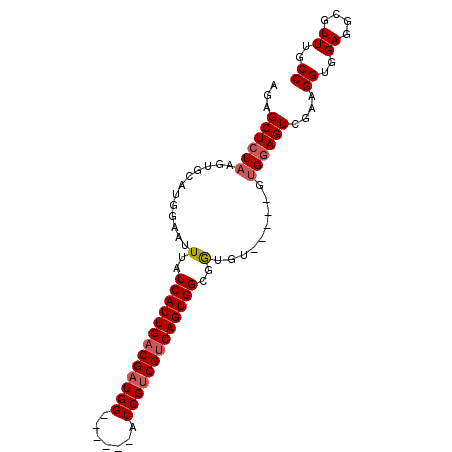
| Location | 13,854,609 – 13,854,724 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -15.59 |
| Energy contribution | -16.70 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13854609 115 + 27905053 UGGAAUUUAUCAUUGACGAUGGUCGACGAUCGUCGUCAGUGGCGGUAA-----AGGAAGUCGGCGGUGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUGGCCAAAGAUCU .........(((((((((((((((...))))))))))))))).(((((-----(.(...(((....)))...).)))))).....((((.(((((.........)))))))))....... ( -36.50) >DroPse_CAF1 184543 88 + 1 UGGAAUUUAUCAUCAUCCAC------------UCAUCGGUGG-------------------GGCAGAGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUG-CUGAAGAUUU ........(((.((..((((------------(....)))))-------------------((((((....((((((((((....))))).....)))).)....))))-)))).))).. ( -22.30) >DroSec_CAF1 125792 109 + 1 UGGAAUUUAUCAUUGACGAUGG------AUCGUCGUCAGUGGCGGUGU-----GUGGAGUCGAAGGUGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUGGCCACAGAUCU .........((((((((((((.------..)))))))))))).(((.(-----((((...((((.(((..(..((((((((....)))))..)))..)..)))..)))).))))).))). ( -37.00) >DroSim_CAF1 131816 109 + 1 UGGAAUUUAUCAUUGACGAUGG------AUCGUCGUCAGUGGCGGUGU-----GUGGAGUCGAAGGUGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUGGCCACAGAUCU .........((((((((((((.------..)))))))))))).(((.(-----((((...((((.(((..(..((((((((....)))))..)))..)..)))..)))).))))).))). ( -37.00) >DroYak_CAF1 136799 114 + 1 UGGAAUUUAUCAUUGCCGUUGG------AUCGUCGUCAGUGGCGAUGGCGGUGGUGGAGUUAGAGGUGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUGGCCACAGAUCU .........((((..(((((..------(((((((....))))))))))))..))))....(((..((..(((...(((((....)))))(((((.........)))))))).))..))) ( -35.70) >DroPer_CAF1 178832 88 + 1 UGGAAUUUAUCAUCAUCCAC------------UCAUCGGUGG-------------------GGCAGAGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUG-CUGAAGAUUU ........(((.((..((((------------(....)))))-------------------((((((....((((((((((....))))).....)))).)....))))-)))).))).. ( -22.30) >consensus UGGAAUUUAUCAUUGACGAUGG______AUCGUCGUCAGUGGCGGUG______GUGGAGUCGGAGGUGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUGGCCAAAGAUCU .........((((((.((((((.......)))))).))))))......................(((.(((.(((((((((....))))).....))))......))).)))........ (-15.59 = -16.70 + 1.11)


| Location | 13,854,649 – 13,854,742 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -14.60 |
| Energy contribution | -14.22 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13854649 93 + 27905053 GGCGGUAA-----AGGAAGUCGGCGGUGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUGGCCAAAGAUCUGGCCAACAUCGCAGAUCA .(((((..-----.....((((((.......))...(((((....))))).......)))).....(((((((......))))))).)))))...... ( -29.40) >DroPse_CAF1 184571 78 + 1 GG-------------------GGCAGAGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUG-CUGAAGAUUUGGCCAACAUCAGAGAUCG ..-------------------((((((.(...).)))))).....((((.(((((..((.((.....))-.))...)))))))))............. ( -17.60) >DroSim_CAF1 131850 93 + 1 GGCGGUGU-----GUGGAGUCGAAGGUGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUGGCCACAGAUCUGGCCAACAUCGCAGAUCA .(((((((-----.....((((((.((....)).))(((((....))))).......))))......((((((......)))))))))))))...... ( -30.90) >DroYak_CAF1 136833 98 + 1 GGCGAUGGCGGUGGUGGAGUUAGAGGUGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUGGCCACAGAUCUGGCCAACAUCGCAGAUCA ...(((.(((((((((..((((....((..(.(((((.....))))).).))..))))..)))...(((((((......)))))))))))))..))). ( -33.80) >DroAna_CAF1 129393 74 + 1 ------------------------GGAGGGGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUGGCCACAGAUCUGGCCAGCAUCGGAUAUCA ------------------------....(..((...(((((....))))).................((((((......))))))))..)........ ( -18.30) >DroPer_CAF1 178860 78 + 1 GG-------------------GGCAGAGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUG-CUGAAGAUUUGGCCAACAUCAGAGAUCG ..-------------------((((((.(...).)))))).....((((.(((((..((.((.....))-.))...)))))))))............. ( -17.60) >consensus GG___________________GGCGGAGGAGGCGUUUGCCAAAAAUGGCACAAAUAACGACAUAAUUUGGCCAAAGAUCUGGCCAACAUCGCAGAUCA ............................(((.(((((((((....))))).....)))).).....(((((((......)))))))..))........ (-14.60 = -14.22 + -0.39)


| Location | 13,854,649 – 13,854,742 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -13.54 |
| Energy contribution | -14.48 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13854649 93 - 27905053 UGAUCUGCGAUGUUGGCCAGAUCUUUGGCCAAAUUAUGUCGUUAUUUGUGCCAUUUUUGGCAAACGCCUCCACCGCCGACUUCCU-----UUACCGCC ......(((...((((((((....)))))))).....((((.......(((((....)))))...((.......)))))).....-----....))). ( -25.20) >DroPse_CAF1 184571 78 - 1 CGAUCUCUGAUGUUGGCCAAAUCUUCAG-CAAAUUAUGUCGUUAUUUGUGCCAUUUUUGGCAAACGCCUCCUCUGCC-------------------CC ...........(((.((((((......(-(((((.........))))))......)))))).)))............-------------------.. ( -13.90) >DroSim_CAF1 131850 93 - 1 UGAUCUGCGAUGUUGGCCAGAUCUGUGGCCAAAUUAUGUCGUUAUUUGUGCCAUUUUUGGCAAACGCCUCCACCUUCGACUCCAC-----ACACCGCC ......(((.(((((((((......))))))......((((.......(((((....)))))..............)))).....-----))).))). ( -24.00) >DroYak_CAF1 136833 98 - 1 UGAUCUGCGAUGUUGGCCAGAUCUGUGGCCAAAUUAUGUCGUUAUUUGUGCCAUUUUUGGCAAACGCCUCCACCUCUAACUCCACCACCGCCAUCGCC ......(((((((((((((......))))))).....(.((((.....(((((....))))))))).).......................)))))). ( -26.60) >DroAna_CAF1 129393 74 - 1 UGAUAUCCGAUGCUGGCCAGAUCUGUGGCCAAAUUAUGUCGUUAUUUGUGCCAUUUUUGGCAAACGCCCCCUCC------------------------ .(((((..(((..((((((......)))))).))))))))........(((((....)))))............------------------------ ( -20.00) >DroPer_CAF1 178860 78 - 1 CGAUCUCUGAUGUUGGCCAAAUCUUCAG-CAAAUUAUGUCGUUAUUUGUGCCAUUUUUGGCAAACGCCUCCUCUGCC-------------------CC ...........(((.((((((......(-(((((.........))))))......)))))).)))............-------------------.. ( -13.90) >consensus UGAUCUCCGAUGUUGGCCAGAUCUGUGGCCAAAUUAUGUCGUUAUUUGUGCCAUUUUUGGCAAACGCCUCCACCGCC___________________CC .......((((((((((((......))))))....)))))).......(((((....))))).................................... (-13.54 = -14.48 + 0.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:35 2006