
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,835,780 – 13,835,884 |
| Length | 104 |
| Max. P | 0.904942 |

| Location | 13,835,780 – 13,835,884 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.96 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -25.25 |
| Energy contribution | -26.03 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
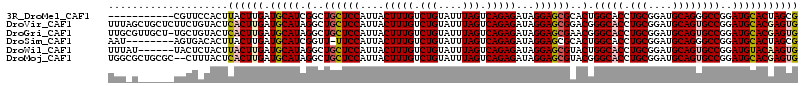
>3R_DroMel_CAF1 13835780 104 + 27905053 CGCUAGUGCAUCCGGCCCUGCAUCCGCAGGUGCCAGUGCGCUCCUAUCUCUGACUAAAUACAGACAAAGUAAUGGAGCAGCCGAUGCAUCAAGUAAGUGGAACG----------- .(((.(((((((.((((((((....))))).))).(.(((((((...((.((.((......)).)).))....))))).))))))))))..)))..........----------- ( -37.50) >DroVir_CAF1 136465 115 + 1 CACUCGUGCAUCCGGCACUGCAUCCGCAGGUGCCCGUCCGCUCCUAUCUCUGACUAAAUACAGACAAAGUAAUGGAGCAGCCUAUGCAUCAAGUGAGUACAGAAGAGCAGCUAAA ((((.((((((..((((((((....))).))))).....(((((...((.((.((......)).)).))....))))).....))))))..)))).................... ( -31.50) >DroGri_CAF1 135825 114 + 1 CACUCGUGCAUCCGGCACUGCAUCCGCAGGUGCCCGUUCGCUCCUAUCUCUGACUAAAUACAGACAAAGUAAUGGAGCAGCCUAUGCAUCAAGUGAGUACAGCA-AGCAACGCAA ((((.((((((..((((((((....))).))))).(((.(((((...((.((.((......)).)).))....))))))))..))))))..))))......((.-......)).. ( -32.90) >DroSim_CAF1 110400 106 + 1 CGCUAGUGCAUCCGGCCCUGCAUCCGCAGGUGCCAGUGCGCUCCUAUCUCUGACUAAAUACAGACAAAGUAAUGGAA-AACCGAUGCAUCAAGUAAGUGUCACU--------AUU (((((((((((..((((((((....))))).))).)))))))......((((........))))....(((.(((..-..))).)))........)))).....--------... ( -31.40) >DroWil_CAF1 101541 109 + 1 CACUUGUACAUCCGGCACUGCAUCCGCAGGUGCCAGUACGCUCCUAUCUCUGACUAAAUACAGACAAAGUAAUGGAGCAGCCUAUGCAUCAAGUAAGUAGAGUA------AUAAA .((((.(((....((((((((....))).)))))..((((((((...((.((.((......)).)).))....))))).((....)).....))).))))))).------..... ( -28.80) >DroMoj_CAF1 159578 113 + 1 CACUCGUGCAUCCGGCACUGCAUCCGCAGGUGCCCGUACGCUCCUAUCUCUGACUAAAUACAGACAAAGUAAUGGAGCAGCCUAUGCAUCAAGUGAGUAAAG--GCGCAGCGCCA .....((((....((((((((....))).))))).((..(((((...((.((.((......)).)).))....))))).((((.(((.((....))))).))--)))).)))).. ( -37.90) >consensus CACUCGUGCAUCCGGCACUGCAUCCGCAGGUGCCAGUACGCUCCUAUCUCUGACUAAAUACAGACAAAGUAAUGGAGCAGCCUAUGCAUCAAGUAAGUACAGCA______C_AAA .((((((((((..((((((((....))).))))).....(((((...((.((.((......)).)).))....))))).....)))))).....))))................. (-25.25 = -26.03 + 0.78)



| Location | 13,835,780 – 13,835,884 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.96 |
| Mean single sequence MFE | -41.68 |
| Consensus MFE | -34.18 |
| Energy contribution | -34.73 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13835780 104 - 27905053 -----------CGUUCCACUUACUUGAUGCAUCGGCUGCUCCAUUACUUUGUCUGUAUUUAGUCAGAGAUAGGAGCGCACUGGCACCUGCGGAUGCAGGGCCGGAUGCACUAGCG -----------.((..((......))..))....((((((((....(((((.(((....))).)))))...)))))((((((((.(((((....)))))))))).)))...))). ( -41.00) >DroVir_CAF1 136465 115 - 1 UUUAGCUGCUCUUCUGUACUCACUUGAUGCAUAGGCUGCUCCAUUACUUUGUCUGUAUUUAGUCAGAGAUAGGAGCGGACGGGCACCUGCGGAUGCAGUGCCGGAUGCACGAGUG ...((.(((......)))))((((((.(((((...(((((((....(((((.(((....))).)))))...))))))).(.(((((.(((....)))))))).)))))))))))) ( -44.70) >DroGri_CAF1 135825 114 - 1 UUGCGUUGCU-UGCUGUACUCACUUGAUGCAUAGGCUGCUCCAUUACUUUGUCUGUAUUUAGUCAGAGAUAGGAGCGAACGGGCACCUGCGGAUGCAGUGCCGGAUGCACGAGUG ..(((.....-)))......((((((.(((((.(..((((((....(((((.(((....))).)))))...))))))..).(((((.(((....))))))))..))))))))))) ( -43.00) >DroSim_CAF1 110400 106 - 1 AAU--------AGUGACACUUACUUGAUGCAUCGGUU-UUCCAUUACUUUGUCUGUAUUUAGUCAGAGAUAGGAGCGCACUGGCACCUGCGGAUGCAGGGCCGGAUGCACUAGCG ..(--------((((.((.........(((.((((..-..))....(((((.(((....))).)))))....))..)))(((((.(((((....)))))))))).)))))))... ( -35.50) >DroWil_CAF1 101541 109 - 1 UUUAU------UACUCUACUUACUUGAUGCAUAGGCUGCUCCAUUACUUUGUCUGUAUUUAGUCAGAGAUAGGAGCGUACUGGCACCUGCGGAUGCAGUGCCGGAUGUACAAGUG .....------.........((((((.(((((.....(((((....(((((.(((....))).)))))...)))))...(((((((.(((....))))))))))))))))))))) ( -40.10) >DroMoj_CAF1 159578 113 - 1 UGGCGCUGCGC--CUUUACUCACUUGAUGCAUAGGCUGCUCCAUUACUUUGUCUGUAUUUAGUCAGAGAUAGGAGCGUACGGGCACCUGCGGAUGCAGUGCCGGAUGCACGAGUG .((((...)))--)......((((((.(((((.....(((((....(((((.(((....))).)))))...)))))...(.(((((.(((....)))))))).)))))))))))) ( -45.80) >consensus UUU_G______UGCUCUACUCACUUGAUGCAUAGGCUGCUCCAUUACUUUGUCUGUAUUUAGUCAGAGAUAGGAGCGCACGGGCACCUGCGGAUGCAGUGCCGGAUGCACGAGUG ....................((((((.(((((.(..((((((....(((((.(((....))).)))))...))))))..).(((((.(((....))))))))..))))))))))) (-34.18 = -34.73 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:30 2006