
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,834,496 – 13,834,723 |
| Length | 227 |
| Max. P | 0.893881 |

| Location | 13,834,496 – 13,834,603 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -18.47 |
| Energy contribution | -18.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13834496 107 - 27905053 UCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAAAUACAGCGGUUUGUAUUUUCCAUUGUUUUCUCUUUUUUAUUUUUUCAAGUAAUUUAUUCUUUCGGCAAAG ((((((.(((((..((((((((..((((...))))...))))))))..)))))....................((((((....)))))).......))))))..... ( -19.80) >DroSec_CAF1 106213 104 - 1 UCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAAAUACAGCGGUUUGUAUUUUCCAUUGUUUUCU---UUUUAUUUUUUCAAGUAAUUUAUUCUUUCGGCAAAG ((((((.(((((..((((((((..((((...))))...))))))))..)))))...............---..((((((....)))))).......))))))..... ( -19.80) >DroSim_CAF1 109134 98 - 1 UCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAAAUACAGCGGUUUGUAUUUUCCAUUGUUUUCU---UU------UUUUAAGUAAUUUAUACUUUCGGCAAAG .....(((((((..((((((((..((((...))))...))))))))..)))))...............---..------....(((((.....)))))...)).... ( -20.00) >DroEre_CAF1 106572 98 - 1 UCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAAAUACAGCGGUUUGUAUUUUCCAUGGUUUUCU---UU------UUUCAAGUAAUUUAUUCUUUCAGCAAAG ..((((.(((((..((((((((..((((...))))...))))))))..)))))))))...........---..------............................ ( -18.40) >DroYak_CAF1 113693 98 - 1 UCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAAAUACAGCGGUUUGUAUUUUCCAUGGUUUUCU---UU------UUUCAAGUAAUUUAUUCUUUCGGCAAAG ((((((.(((((..((((((((..((((...))))...))))))))..)))))......(((......---..------..)))............))))))..... ( -18.70) >DroAna_CAF1 104371 93 - 1 UCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAGAUACAGCGGUUUGUAUUUUCUAUGUUUUU--------------GUUAUGUAAUUUCAUAAUUUAGCAAAG ..((((.(((((..((((((((..((((...))))...))))))))..)))))))))..((((...--------------((((((......))))))..))))... ( -25.20) >consensus UCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAAAUACAGCGGUUUGUAUUUUCCAUGGUUUUCU___UU______UUUCAAGUAAUUUAUUCUUUCGGCAAAG ..((((.(((((..((((((((..((((...))))...))))))))..))))))))).................................................. (-18.47 = -18.47 + -0.00)


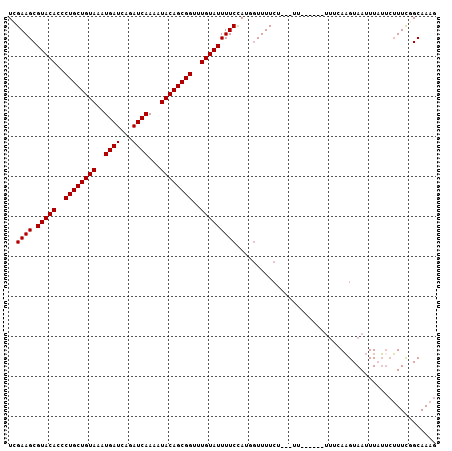
| Location | 13,834,529 – 13,834,643 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.46 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -23.18 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13834529 114 + 27905053 AAAAAG------AGAAAACAAUGGAAAAUACAAACCGCUGUAUUUUGAUCUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAG ......------.(((.....(((....((((..(.((((((...((((...))))..)))))).)..)))).((.(((....)))...)).))).....))).(((((......))))) ( -26.00) >DroVir_CAF1 135080 115 + 1 AAAAGUACAAAAU-----GAAAAAAAAAUUCAAACCGCUGUAUUUUGAUCUGAUCACUUACAGCAGGGUGUACGCUUCGACCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAG ....(((((...(-----(((.......))))..((((((((...((((...))))..)))))).)).)))))((.(((....)))...)).............(((((......))))) ( -24.90) >DroGri_CAF1 134540 120 + 1 AAAAGUGAAAAAAAAAAAAAAUAGAAAAUACAAACCGCUGUAUUUUGAUCUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAG ..(((((((...........................((((((...((((...))))..)))))).(((((((....(((....)))))))))).......)))))))((......))... ( -26.40) >DroWil_CAF1 100262 111 + 1 AAC---------AAAAAAAUAUAGAAAAUACAAACCGCUGUAUCUUGAUUUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUACGCUUGGAAUUCGCCAAG ...---------........................((((((...((((...))))..)))))).(((((((....(((....))))))))))...........(((((......))))) ( -23.30) >DroMoj_CAF1 158246 115 + 1 GAAAAUGCAAAUU-----AAAUAGAAUAUACAAACCGCUGUAUCUUGAUCUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAG .............-----.....((((.........((((((...((((...))))..)))))).(((((((....(((....))))))))))......)))).(((((......))))) ( -25.50) >DroAna_CAF1 104398 106 + 1 --------------AAAAACAUAGAAAAUACAAACCGCUGUAUCUUGAUCUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAG --------------.........(((..........((((((...((((...))))..)))))).(((((((....(((....)))))))))).......))).(((((......))))) ( -24.50) >consensus AAAA_U______A_AAAAAAAUAGAAAAUACAAACCGCUGUAUCUUGAUCUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAG ....................................((((((...((((...))))..)))))).(((((((....(((....))))))))))...........(((((......))))) (-23.18 = -23.18 + 0.00)

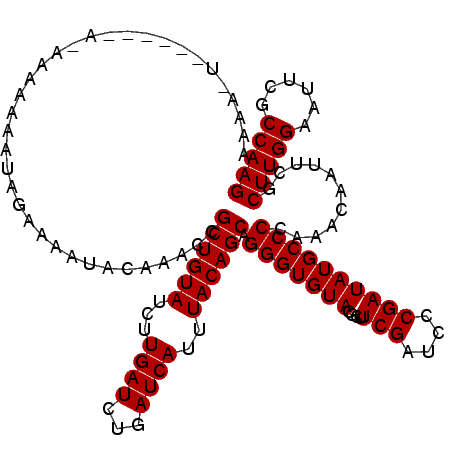
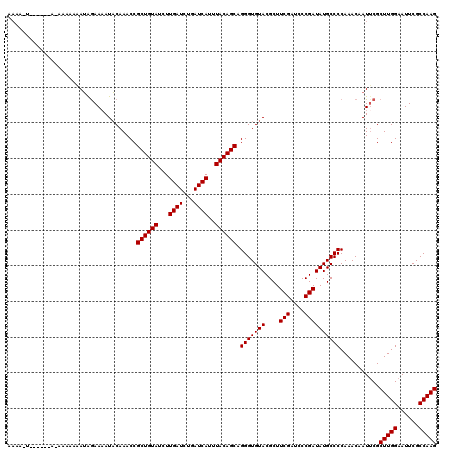
| Location | 13,834,529 – 13,834,643 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.46 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -27.64 |
| Energy contribution | -27.83 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13834529 114 - 27905053 CUUGGCGAAUUCCAAGCGAAUUGUUUGGGGCAUAUCGGGAUCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAAAUACAGCGGUUUGUAUUUUCCAUUGUUUUCU------CUUUUU ((((..(..((((((((.....))))))))..)..))))...((((.(((((..((((((((..((((...))))...))))))))..)))))))))...........------...... ( -28.50) >DroVir_CAF1 135080 115 - 1 CUUGGCGAAUUCCAAGCGAAUUGUUUGGGGCAUAUCGGGGUCGAAGCGUACACCCUGCUGUAAGUGAUCAGAUCAAAAUACAGCGGUUUGAAUUUUUUUUUC-----AUUUUGUACUUUU ....((...((((((((.....))))))))....(((....))).))(((((..((((((((..((((...))))...))))))))..((((.......)))-----)...))))).... ( -28.70) >DroGri_CAF1 134540 120 - 1 CUUGGCGAAUUCCAAGCGAAUUGUUUGGGGCAUAUCGGGAUCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAAAUACAGCGGUUUGUAUUUUCUAUUUUUUUUUUUUUUCACUUUU ((((..(..((((((((.....))))))))..)..))))...((((.(((((..((((((((..((((...))))...))))))))..)))))))))....................... ( -28.90) >DroWil_CAF1 100262 111 - 1 CUUGGCGAAUUCCAAGCGUAUUGUUUGGGGCAUAUCGGGAUCGAAGCGUACACCCUGCUGUAAAUGAUCAAAUCAAGAUACAGCGGUUUGUAUUUUCUAUAUUUUUUU---------GUU ((((..(..((((((((.....))))))))..)..))))...((((.(((((..((((((((..((((...))))...))))))))..)))))))))...........---------... ( -28.90) >DroMoj_CAF1 158246 115 - 1 CUUGGCGAAUUCCAAGCGAAUUGUUUGGGGCAUAUCGGGAUCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAGAUACAGCGGUUUGUAUAUUCUAUUU-----AAUUUGCAUUUUC .((((......))))((((((((..((((((...(((....))).))(((((..((((((((..((((...))))...))))))))..)))))..))))..)-----)))))))...... ( -32.90) >DroAna_CAF1 104398 106 - 1 CUUGGCGAAUUCCAAGCGAAUUGUUUGGGGCAUAUCGGGAUCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAGAUACAGCGGUUUGUAUUUUCUAUGUUUUU-------------- ((((..(..((((((((.....))))))))..)..))))...((((.(((((..((((((((..((((...))))...))))))))..))))))))).........-------------- ( -28.90) >consensus CUUGGCGAAUUCCAAGCGAAUUGUUUGGGGCAUAUCGGGAUCGAAGCGUACACCCUGCUGUAAAUGAUCAGAUCAAAAUACAGCGGUUUGUAUUUUCUAUUUUUUU_U______A_UUUU ((((..(..((((((((.....))))))))..)..))))...((((.(((((..((((((((..((((...))))...))))))))..)))))))))....................... (-27.64 = -27.83 + 0.19)



| Location | 13,834,563 – 13,834,683 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.39 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -24.84 |
| Energy contribution | -24.70 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13834563 120 + 27905053 UAUUUUGAUCUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUA ....(((..(((.....((((....(((((((....(((....))))))))))...........(((((......))))).))))(((....)))...............)))..))).. ( -25.10) >DroVir_CAF1 135115 120 + 1 UAUUUUGAUCUGAUCACUUACAGCAGGGUGUACGCUUCGACCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUA ....(((..(((.....((((....(((((((....(((....))))))))))...........(((((......))))).))))(((....)))...............)))..))).. ( -25.00) >DroWil_CAF1 100293 120 + 1 UAUCUUGAUUUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUACGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUA ......................((.(((((((....(((....)))))))))).......(((.(((((......))))).))).(((....))).))...................... ( -24.80) >DroMoj_CAF1 158281 120 + 1 UAUCUUGAUCUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUA ....(((..(((.....((((....(((((((....(((....))))))))))...........(((((......))))).))))(((....)))...............)))..))).. ( -25.00) >DroAna_CAF1 104424 120 + 1 UAUCUUGAUCUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUA ....(((..(((.....((((....(((((((....(((....))))))))))...........(((((......))))).))))(((....)))...............)))..))).. ( -25.00) >DroPer_CAF1 155009 120 + 1 UAUUUUGAUCUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUA ....(((..(((.....((((....(((((((....(((....))))))))))...........(((((......))))).))))(((....)))...............)))..))).. ( -25.10) >consensus UAUCUUGAUCUGAUCAUUUACAGCAGGGUGUACGCUUCGAUCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUA ....(((..(((.....((((....(((((((....(((....))))))))))...........(((((......))))).))))(((....)))...............)))..))).. (-24.84 = -24.70 + -0.14)



| Location | 13,834,603 – 13,834,723 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.12 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13834603 120 + 27905053 UCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUACAGCGACAACAGCCUCACAUCCUGCAUUGAUGCACCCACU ........(((.............(((((......))))).....(((....))).))).............((.((((.(((.((.............))))).))))..))....... ( -19.12) >DroVir_CAF1 135155 120 + 1 CCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUACAGCGACAACAGCCUCACAUCCUGCAUUGAUGCACCCACU ........(((.............(((((......))))).....(((....))).))).............((.((((.(((.((.............))))).))))..))....... ( -19.12) >DroGri_CAF1 134620 120 + 1 UCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUACAGCGACAACAGCCUCACAUCCUGCAUUGAUGCACCCUCU ........(((.............(((((......))))).....(((....))).))).............((.((((.(((.((.............))))).))))..))....... ( -19.12) >DroWil_CAF1 100333 120 + 1 UCCCGAUAUGCCCCAAACAAUACGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUACAGCGACAACAGCCUCACAUCCUGCAUUGAUGCACCCACU ........(((.........(((.(((((......))))).))).(((....))).))).............((.((((.(((.((.............))))).))))..))....... ( -22.02) >DroAna_CAF1 104464 120 + 1 UCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUACAGCGACAACAGCCUCACAUCCUGCAUUGAUGCACCCACU ........(((.............(((((......))))).....(((....))).))).............((.((((.(((.((.............))))).))))..))....... ( -19.12) >DroPer_CAF1 155049 120 + 1 UCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUACAGCGACAACAGCCUCACAUCCUGCAUUGAUGCACCCACU ........(((.............(((((......))))).....(((....))).))).............((.((((.(((.((.............))))).))))..))....... ( -19.12) >consensus UCCCGAUAUGCCCCAAACAAUUCGCUUGGAAUUCGCCAAGAGUAACACGAAAGUGAGCAAACCCAAACCACAGCCCAAUACAGCGACAACAGCCUCACAUCCUGCAUUGAUGCACCCACU ........(((.............(((((......))))).....(((....))).))).............((.((((.(((.((.............))))).))))..))....... (-19.12 = -19.12 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:28 2006