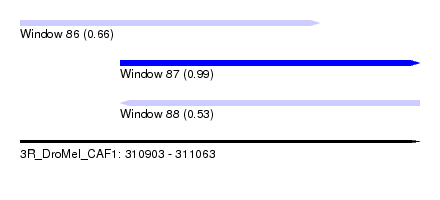
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 310,903 – 311,063 |
| Length | 160 |
| Max. P | 0.991423 |

| Location | 310,903 – 311,023 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -52.73 |
| Consensus MFE | -49.29 |
| Energy contribution | -49.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 310903 120 + 27905053 ACGGCGGGGAUUCUCUGACGGCGGAGCACGAGUCGGCGGCGGCUGAGGCUCAGGCAUUGGCCGAAUUGGUGGCCGGCGAAAAGCCGGGCGCUUCGGCCGGAACUAACUCCACUGAAGACU .(((.((((..((.(((((..((.....)).))))).))((((((((((...(((....)))........(.(((((.....))))).))))))))))).......)))).)))...... ( -54.70) >DroSec_CAF1 47845 120 + 1 ACGGCGGGGAUUCUCUCACGGCGGAGCACGAGUCGGCGGCGGCUGAGGCUCAGGCAUUGGCCGAAUUGGUGGCCGGCGAAAAGCCGGGUGCUUCGGCCGGAACUAAUUCCGCUGAAGACU .(((((((.(((...((.(((((((((((..((((.((.((((..(.((....)).)..))))...)).))))((((.....)))).))))))).))))))...))))))))))...... ( -56.00) >DroSim_CAF1 48701 120 + 1 ACGGCGGGGAUUCUCUCACGGCGGAGCACGAGUCGGCGGCGGCUGAGGCUCAGGCAUUGGCCGAAUUGGUGGCCGGCGAAAAGCCGGGCGCUUCGGCCGGAACUAAUUCCGCUGAAGACU .((.(((((.....))).)).)).......((((..(((((((((((((...(((....)))........(.(((((.....))))).))))))))))((((....))))))))..)))) ( -57.60) >DroYak_CAF1 51123 117 + 1 ACGGCGGGGAUUCUCUGAUGGCGGAACACGAGUCGGU---GGCUGAGGCUCAGGCAUUGGCUGAAUUGGUGGCCGGCGAAAAGAUGGGCGCUUCGGUAGGGACUGAUUCCGCUGAAGACU .((((((((..((((((..((((...(((..((((((---.((..(...((((.(...).)))).)..)).)))))).....).))..))))....))))))....))))))))...... ( -42.60) >consensus ACGGCGGGGAUUCUCUCACGGCGGAGCACGAGUCGGCGGCGGCUGAGGCUCAGGCAUUGGCCGAAUUGGUGGCCGGCGAAAAGCCGGGCGCUUCGGCCGGAACUAAUUCCGCUGAAGACU .((((((((...........((.((((...(((((....)))))...))))..)).((((((((...((((.(((((.....))))).))))))))))))......))))))))...... (-49.29 = -49.60 + 0.31)

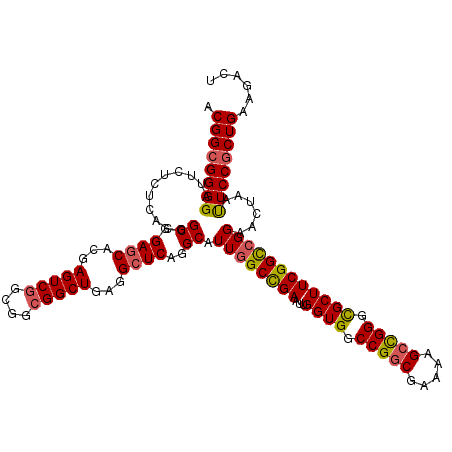
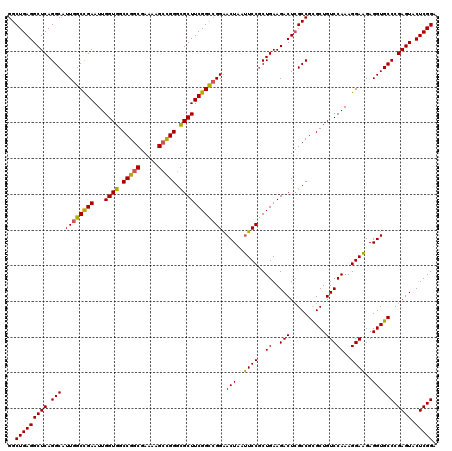
| Location | 310,943 – 311,063 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -56.62 |
| Consensus MFE | -52.72 |
| Energy contribution | -52.10 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 310943 120 + 27905053 GGCUGAGGCUCAGGCAUUGGCCGAAUUGGUGGCCGGCGAAAAGCCGGGCGCUUCGGCCGGAACUAACUCCACUGAAGACUCGCCGCGCUGUCCAAAGGAGGAGGUGCCCGAGUACUCGGA (((.(((..((((.....((((((...((((.(((((.....))))).))))))))))(((......))).))))...))))))(((((.(((......))))))))((((....)))). ( -59.80) >DroSec_CAF1 47885 120 + 1 GGCUGAGGCUCAGGCAUUGGCCGAAUUGGUGGCCGGCGAAAAGCCGGGUGCUUCGGCCGGAACUAAUUCCGCUGAAGACUCGCCGCGCUGUCCAAAGGAAGAGGUGCCCGAGUACUCGGA (((.(((.((..(((...((((((...((((.(((((.....))))).))))))))))((((....)))))))..)).))))))(((((.(((...)))...)))))((((....)))). ( -58.20) >DroSim_CAF1 48741 120 + 1 GGCUGAGGCUCAGGCAUUGGCCGAAUUGGUGGCCGGCGAAAAGCCGGGCGCUUCGGCCGGAACUAAUUCCGCUGAAGACUGGCCGUGCUGUCCAAAGGAAGAGGUGCCCGAGUACUCGGA ..(((((((((.(((.((((((((...((((.(((((.....))))).)))))))))))).(((..((((..((..(((.(((...))))))))..))))..)))))).)))).))))). ( -57.90) >DroYak_CAF1 51160 120 + 1 GGCUGAGGCUCAGGCAUUGGCUGAAUUGGUGGCCGGCGAAAAGAUGGGCGCUUCGGUAGGGACUGAUUCCGCUGAAGACUCGCCGCGCUGUCCAAAGGAAGAGGUGCCCGAGUACUCGGA ..(((((((((.((((((..((...((((((((((((((.....(.((((..(((((....)))))...)))).)....)))))).)))).))))....)).)))))).)))).))))). ( -50.60) >consensus GGCUGAGGCUCAGGCAUUGGCCGAAUUGGUGGCCGGCGAAAAGCCGGGCGCUUCGGCCGGAACUAAUUCCGCUGAAGACUCGCCGCGCUGUCCAAAGGAAGAGGUGCCCGAGUACUCGGA ..(((((((((.(((.((((((((...((((.(((((.....))))).)))))))))))).(((..((((..((..(((..........)))))..))))..)))))).)))).))))). (-52.72 = -52.10 + -0.62)

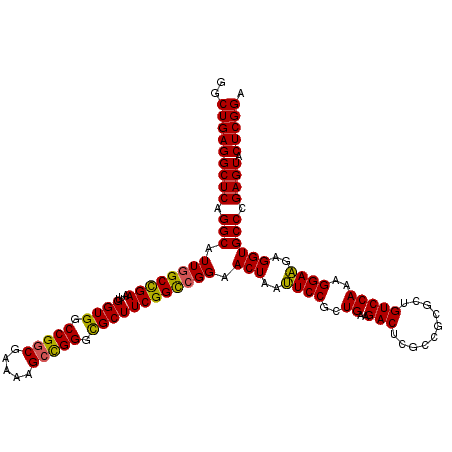
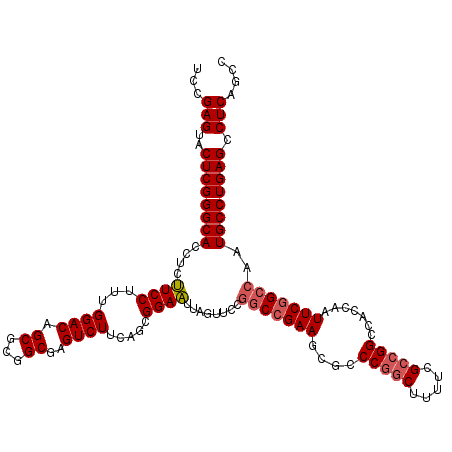
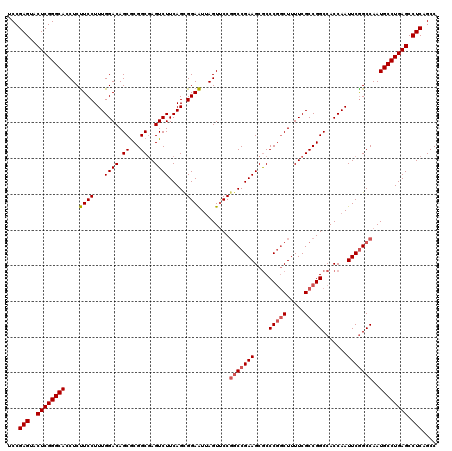
| Location | 310,943 – 311,063 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -44.57 |
| Consensus MFE | -39.10 |
| Energy contribution | -39.98 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 310943 120 - 27905053 UCCGAGUACUCGGGCACCUCCUCCUUUGGACAGCGCGGCGAGUCUUCAGUGGAGUUAGUUCCGGCCGAAGCGCCCGGCUUUUCGCCGGCCACCAAUUCGGCCAAUGCCUGAGCCUCAGCC ...(((..((((((((.((((..((..((((.((...))..))))..)).))))........((((((((.(.(((((.....))))).).)...)))))))..)))))))).))).... ( -49.30) >DroSec_CAF1 47885 120 - 1 UCCGAGUACUCGGGCACCUCUUCCUUUGGACAGCGCGGCGAGUCUUCAGCGGAAUUAGUUCCGGCCGAAGCACCCGGCUUUUCGCCGGCCACCAAUUCGGCCAAUGCCUGAGCCUCAGCC ...(((..((((((((.......((..((((.((...))..))))..)).((((....))))(((((((....(((((.....))))).......)))))))..)))))))).))).... ( -46.20) >DroSim_CAF1 48741 120 - 1 UCCGAGUACUCGGGCACCUCUUCCUUUGGACAGCACGGCCAGUCUUCAGCGGAAUUAGUUCCGGCCGAAGCGCCCGGCUUUUCGCCGGCCACCAAUUCGGCCAAUGCCUGAGCCUCAGCC ...(((..((((((((((.........)).......((((.(.((((.((((((....)))).)).)))))(.(((((.....))))).)........))))..)))))))).))).... ( -46.30) >DroYak_CAF1 51160 120 - 1 UCCGAGUACUCGGGCACCUCUUCCUUUGGACAGCGCGGCGAGUCUUCAGCGGAAUCAGUCCCUACCGAAGCGCCCAUCUUUUCGCCGGCCACCAAUUCAGCCAAUGCCUGAGCCUCAGCC ...(((..((((((((.........((((...((.(((((((......(((...((.((....)).))..))).......)))))))))..)))).........)))))))).))).... ( -36.49) >consensus UCCGAGUACUCGGGCACCUCUUCCUUUGGACAGCGCGGCGAGUCUUCAGCGGAAUUAGUUCCGGCCGAAGCGCCCGGCUUUUCGCCGGCCACCAAUUCGGCCAAUGCCUGAGCCUCAGCC ...(((..((((((((....((((...((((.((...))..)))).....))))........(((((((....(((((.....))))).......)))))))..)))))))).))).... (-39.10 = -39.98 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:31 2006