
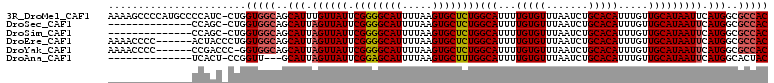
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,820,251 – 13,820,394 |
| Length | 143 |
| Max. P | 0.987305 |

| Location | 13,820,251 – 13,820,360 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.05 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -27.21 |
| Energy contribution | -27.60 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
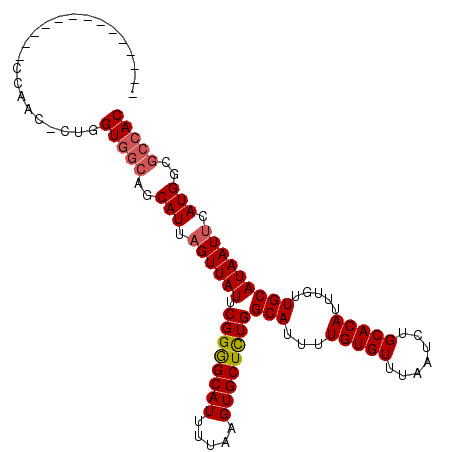
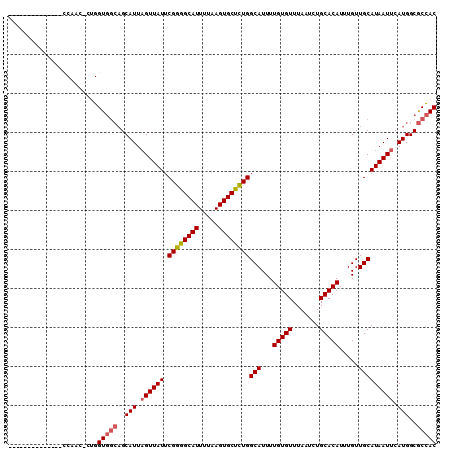
>3R_DroMel_CAF1 13820251 109 + 27905053 AAAAGCCCCAUGCCCCAUC-CUGGUGGCAGCAUUUGUUAUUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCAC ....((.(((((...(((.-...)))((((((..(((....((((((((.....))))))))(((..(((....)))..))))))..))))))......))))).))... ( -34.60) >DroSec_CAF1 92144 95 + 1 --------------CCAGC-CUGGUGGCAGCAUUAGUUAUUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCAC --------------.....-...(((((..(((.((((((.((((((((.....))))))))(((...(((((.......))))).....))))))))).)))..))))) ( -29.80) >DroSim_CAF1 94202 95 + 1 --------------CCAGC-CUGGUGGCAGCAUUAGUUAUUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCAC --------------.....-...(((((..(((.((((((.((((((((.....))))))))(((...(((((.......))))).....))))))))).)))..))))) ( -29.80) >DroEre_CAF1 90062 104 + 1 AAAACCCC------ACUACCCUGGUGGCAGCAUUAGUUAUUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCAC ........------.........(((((..(((.((((((.((((((((.....))))))))(((...(((((.......))))).....))))))))).)))..))))) ( -29.80) >DroYak_CAF1 99383 103 + 1 AAAACCCC------CCGACCC-GGUGGCAGCAUUAGUUAUUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCAC ........------.......-.(((((..(((.((((((.((((((((.....))))))))(((...(((((.......))))).....))))))))).)))..))))) ( -29.80) >DroAna_CAF1 91437 92 + 1 --------------UCACU-CCGGUU---GCAUUAGUUAUUCGGAGCAUUUUAAGUGCUUUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCACUAC --------------...((-((((..---((....))...)))))).......((((((.(((((...(((((.......))))).....)))......)).)))))).. ( -23.10) >consensus ______________CCAAC_CUGGUGGCAGCAUUAGUUAUUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCAC .......................(((((..(((.((((((.((((((((.....))))))))(((...(((((.......))))).....))))))))).)))..))))) (-27.21 = -27.60 + 0.39)
| Location | 13,820,289 – 13,820,394 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -22.79 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
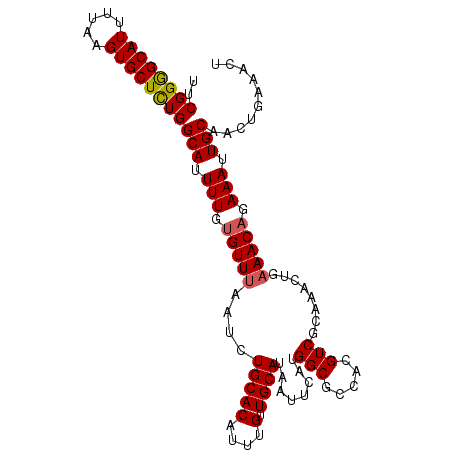
>3R_DroMel_CAF1 13820289 105 + 27905053 UUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCACGUCGCAAACUGAAACAGAAAUUGCAACUGAAACU ..((((((((.....))))))))(((.(((.(((((....(((((....).)))).........(((......)))......))))).))).))).......... ( -25.70) >DroSec_CAF1 92168 105 + 1 UUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCACGUCGCAAACUGAAACAAAAAUUGCCACUGAAACU (((((((((.(((..((((...((((...(((((.......)))))..))))))))..((((..(((......)))....))))....))).)))).)))))... ( -26.50) >DroSim_CAF1 94226 105 + 1 UUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCACGUCGCAAACUGAAACAGAAAUUGCCACUGAAACU ..((((((((.....))))))))(((...(((((.......))))).....)))....((((((((((......))...(((...))).....)))).))))... ( -28.30) >DroEre_CAF1 90095 105 + 1 UUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCACGUCGCAAACUGAAACAGAAAUUGCAACUGAAACU ..((((((((.....))))))))(((.(((.(((((....(((((....).)))).........(((......)))......))))).))).))).......... ( -25.70) >DroYak_CAF1 99415 105 + 1 UUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCACGUCGCAAACUGAAACAGAAAUUGCAACUGAAACU ..((((((((.....))))))))(((.(((.(((((....(((((....).)))).........(((......)))......))))).))).))).......... ( -25.70) >DroAna_CAF1 91458 104 + 1 UUCGGAGCAUUUUAAGUGCUUUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCACUACGUCGCAAACUGCAACUGAAAUUGC-ACUGCCACU ...(((((((.....)))))))((((...(((((.......))))).....((((...((((.(((.....)))((.....))...))))..)))-).))))... ( -25.50) >consensus UUCGGGGCAUUUUAAGUGCUCUGGCAUUUUGUGUUUAAUCUGCACAUUUGUUGCAUAAUUCAUGGCGCCACGUCGCAAACUGAAACAGAAAUUGCAACUGAAACU ..((((((((.....))))))))(((.(((.(((((....(((((....).))))........(((.....)))........))))).))).))).......... (-22.79 = -22.85 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:14 2006