| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,943,903 – 1,943,995 |
| Length | 92 |
| Max. P | 0.532179 |

| Location | 1,943,903 – 1,943,995 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.85 |
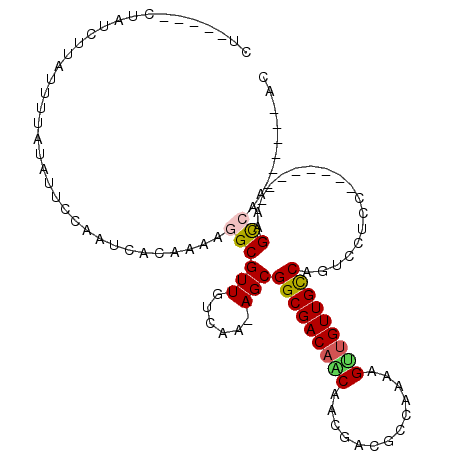
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.24 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
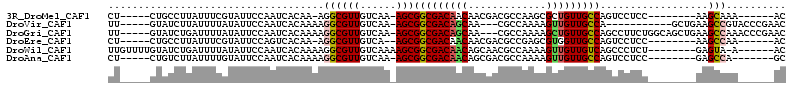
>3R_DroMel_CAF1 1943903 92 - 27905053 CU-----CUGCCUUAUUUCGUAUUCCAAUCACAA-AGGCGUUGUCAA-AGCGGCGACAACAACGACGCCAAGCGCUGUUGCCAGUCCUCC--------AAGCAAA------AC ..-----..(((((...................)-))))((((((..-...))))))..(((((.(((...))).)))))..........--------.......------.. ( -18.01) >DroVir_CAF1 121359 93 - 1 UU-----GUAUCUUAUUUUAUAUUCCAAUCACAAAAGGCGUUGUCAA-AGCGGCGACAGCAA---CGCCAAAAGUUGUUGCCA-----------GCUGAAGCCGUACCCGAAC ..-----.............................(((....(((.-.(((((((((((..---........))))))))).-----------))))).))).......... ( -21.80) >DroGri_CAF1 103745 104 - 1 UU-----GUAUCUGAUUUUAUAUUCCAAUCACAAAAGGCGUUGUCAA-AGCGGCGACAGCAA---CGCCAAAAGCUGUUGCCAGCCUUCUGGCAGCUGAAGCCAAACCCGAAC ((-----((((..((........))..)).))))..(((....(((.-.(((((((((((..---........))))))))).(((....))).))))).))).......... ( -30.30) >DroEre_CAF1 104311 91 - 1 CU-----CUGCCUUAUUUCGUAUUCCAGUCACAA-AGGCGUUGUCA--AGCGGCGACAACAACGACGCCGAGCGUGGUUGCCAGUCCUCC--------AAGCCAA------AC ..-----...........................-.(((((((((.--...))))))..((((.((((...)))).))))..........--------..)))..------.. ( -22.10) >DroWil_CAF1 102518 98 - 1 UUGUUUUGUAUCUGAUUUUAUAUUCCAAUCACAAAAGGCGUUGUCAAAAGCGGCGACAACAGCAACGCCAAAAGUUGUUGUCAGCCCUCU--------GAGUA-A------AC ...((((((((..((........))..)).))))))(((((((((......)))))).((((((((.......))))))))..)))....--------.....-.------.. ( -23.20) >DroAna_CAF1 86680 92 - 1 CU-----CUGUCUUAUUUUGUAUUCCAAUCACAAAAGGCGUUGUCAA-AGCGGCGACAACAGCGACGCCAAAAGUUGUUGCCAGUCCUCC--------GAGCCA-------GC ..-----(((.(((.((((((.........))))))(((((((((..-......))))))((((((.......)))))))))........--------))).))-------). ( -23.30) >consensus CU_____CUAUCUUAUUUUAUAUUCCAAUCACAAAAGGCGUUGUCAA_AGCGGCGACAACAACGACGCCAAAAGUUGUUGCCAGUCCUCC________AAGCCAA______AC ....................................((((((......)))(((((((((.............)))))))))..................))).......... (-16.12 = -16.24 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:19 2006