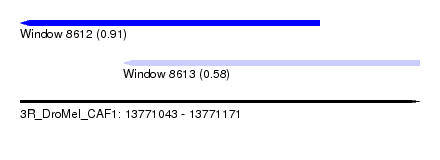
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,771,043 – 13,771,171 |
| Length | 128 |
| Max. P | 0.910183 |

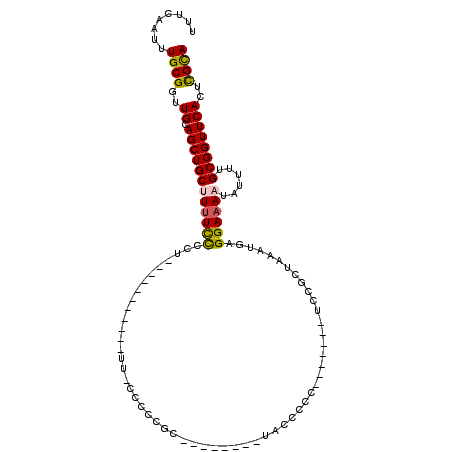
| Location | 13,771,043 – 13,771,139 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.20 |
| Mean single sequence MFE | -17.00 |
| Consensus MFE | -11.61 |
| Energy contribution | -11.59 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13771043 96 - 27905053 UUUCCCCUCCCUCUCUUCAAUCCUC-------ACCACUAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCAAGAUUAUGGCUCAAGAUUGC-AAUCCCUUAACUA ....................(((((-------(........))))))....(((((((((.....)))))))))....((........))-............. ( -16.70) >DroSec_CAF1 43602 88 - 1 UUCCCCCCGC--------UACCCUA-------UACGCCAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCAAGAUUAUGGCCCAAGAUUGC-AAUCCCUUAACUA ..........--------.......-------.........(((((.....((((((.((((...((....))....))))))))))...-....))))).... ( -14.30) >DroSim_CAF1 42492 88 - 1 UUCCCCCCGC--------UACCCUA-------UCCGCCAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCAAGAUUAUGGCCCAAGAUUGC-AAUCCCUUAACUA ..........--------.......-------.........(((((.....((((((.((((...((....))....))))))))))...-....))))).... ( -14.30) >DroEre_CAF1 42321 86 - 1 ---CCCCCAC--------UACCCCC-------UCCGCUAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCAAGAUUAUGGCCCAAGAUUGCCAAUCCCUUAACUA ---.......--------.......-------.........(((((.....(((((((((.....)))))))))..((((........))))...))))).... ( -17.50) >DroYak_CAF1 49980 93 - 1 ---CCCCCUC--------GACCCCCCCUACUAUCCGCUAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCAAGAUUACGGCCCAAGAUUGCCAAUCCCUCAACUA ---.......--------.......................(((((.....(((((((((.....)))))))))...(((........)))....))))).... ( -19.00) >DroAna_CAF1 45065 72 - 1 ---GCCCU--------------------------GGCUAAAUGGGGAA-CUA-UUUGCGGUUCACAUGUAAGAUUAUGGCCCAUGAUUGC-CAAUACCUAACUA ---....(--------------------------(((...((((((((-((.-.....))))).((((......)))).)))))....))-))........... ( -20.20) >consensus ___CCCCCGC________UACCCUC_______UCCGCUAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCAAGAUUAUGGCCCAAGAUUGC_AAUCCCUUAACUA .........................................(((((.....((((((.((((...((....))....))))))))))........))))).... (-11.61 = -11.59 + -0.03)

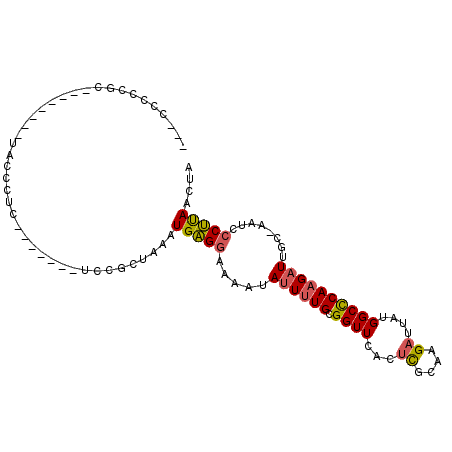
| Location | 13,771,076 – 13,771,171 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -17.67 |
| Consensus MFE | -12.35 |
| Energy contribution | -12.30 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13771076 95 - 27905053 UUUCAAUUUGCGGUUGCAGCUGCUUUUUCCCU-----------UUUCCCCUCCCUCUCUUCAAUCCUC-------ACCACUAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCA ........((((..((.((((((.........-----------....................(((((-------(........)))))).((....))))))))))..)))) ( -19.10) >DroSec_CAF1 43635 87 - 1 UUUCAAUUUGCGGUUGCAGCUGCUUUUCCCCU-----------UUCCCCCCGC--------UACCCUA-------UACGCCAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCA ........((((..((.((((((........(-----------(((((...((--------.......-------...)).....).))))).......))))))))..)))) ( -16.76) >DroSim_CAF1 42525 87 - 1 UUUCAAUUUGCGGUUGCAGCUGCUUUUCCCCU-----------UUCCCCCCGC--------UACCCUA-------UCCGCCAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCA ........((((..((.((((((........(-----------(((((...((--------.......-------...)).....).))))).......))))))))..)))) ( -16.76) >DroEre_CAF1 42355 84 - 1 UUUCAAUUUGCGGUUGCAGCUGCUUUUCCCUU--------------CCCCCAC--------UACCCCC-------UCCGCUAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCA ........((((..((.((((((.........--------------.......--------.....((-------((........))))(((....)))))))))))..)))) ( -16.30) >DroYak_CAF1 50014 91 - 1 UUUCAAUUUGCGGUUGCAGCUGCCUUUCCGUU--------------CCCCCUC--------GACCCCCCCUACUAUCCGCUAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCA ........((((..((.((((((.....((..--------------......)--------).............(((.(.....).))).........))))))))..)))) ( -16.90) >DroPer_CAF1 58806 97 - 1 UUUCAAUUUGCGGUUGCAGCUGCCUGUCUGCCUGUCUGCCGCCUUGCCCUUGC--------CAGCUCC-------ACUCCUAAAUGAGGAAAAUAU-UUGCGGUUCACGUGUA .........(((((....))))).....(((.(((..(((((((.((....))--------.))....-------..((((.....))))......-..)))))..))).))) ( -20.20) >consensus UUUCAAUUUGCGGUUGCAGCUGCUUUUCCCCU___________UU_CCCCCGC________UACCCCC_______UCCGCUAAAUGAGGAAAAUAUUUUGCGGUUCACUCGCA ........((((..((.((((((((((((..........................................................))))))......))))))))..)))) (-12.35 = -12.30 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:55 2006