| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,941,775 – 1,941,884 |
| Length | 109 |
| Max. P | 0.827077 |

| Location | 1,941,775 – 1,941,884 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -22.60 |
| Energy contribution | -22.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
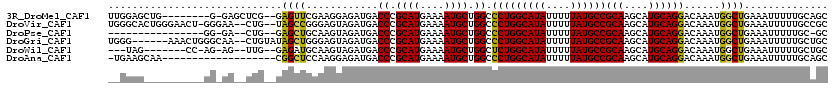
>3R_DroMel_CAF1 1941775 109 + 27905053 GCUGCAAAAAUUUCAGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUCCUUCGAACUC--CGAGCUC-C--------CAGCUCCAA ((((.((....))))))....((.((((((((((.((((((((....))))))...)).))))......)))))).))..............--.((((..-.--------..))))... ( -30.30) >DroVir_CAF1 118508 115 + 1 GCGGCAAAAAUUUCAGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUACUCCCGGCUA--CAG--UUCCC-AGUUCCCAGUGCCCA (.((((........((((....((((((((....)))((((((....)))))))))))..((((....))))(((.........)))))))(--(..--.....-.))......))))). ( -33.50) >DroPse_CAF1 83555 98 + 1 GC-GCAAAAAUUUCAGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUACUUGCAGCUC--CAG--UC-CC---------------- ((-((((........(((....((((((((....)))((((((....)))))))))))..((((....)))).))).......)))).))..--...--..-..---------------- ( -27.36) >DroGri_CAF1 101333 112 + 1 GCAGCAAAAAUUUCAGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUACUCCCAGCUAUACAG--UUGCCCAGUUU------CCCA (((((.................((((((((....)))((((((....)))))))))))..((((....))))(((.........))).........)--)))).......------.... ( -29.70) >DroWil_CAF1 99785 104 + 1 GCAGCAAAAAUUUCAGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGAGCCAGCAUUUUCAUGCGGGUCAUCUACUUGCAUCUC--CAA--CU-CU-GG-------CUA--- ((((((.......(((....))).......))).)))((((((....))))))..((((((.......((((((((.....))))))))...--...--..-))-))-------)).--- ( -30.00) >DroAna_CAF1 84603 100 + 1 GCUGCAAAAAUUUCAGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUCCUUGGAGCCG-------------------UUGCUUCA- (..((((...((((((.....((.((((((((((.((((((((....))))))...)).))))......)))))).)).....))))))...-------------------))))..).- ( -30.70) >consensus GCAGCAAAAAUUUCAGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUACUUCCAGCUC__CAG__UU_CC__________CUCC__ .....................((.((((((((((.((((((((....))))))...)).))))......)))))).)).......................................... (-22.60 = -22.60 + -0.00)


| Location | 1,941,775 – 1,941,884 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -23.69 |
| Energy contribution | -23.33 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1941775 109 - 27905053 UUGGAGCUG--------G-GAGCUCG--GAGUUCGAAGGAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCUGAAAUUUUUGCAGC ..((.((((--------(-(...(((--...(((....))).))))))((((....)))).))))).((((((....))))))(((((....(((..(....).))).....)))))... ( -33.20) >DroVir_CAF1 118508 115 - 1 UGGGCACUGGGAACU-GGGAA--CUG--UAGCCGGGAGUAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCUGAAAUUUUUGCCGC ..(((((..(...).-.)...--...--((((((((.........)))((((....))))((..(((((((((....))))))(((....)))))).))...))))).......)))).. ( -35.90) >DroPse_CAF1 83555 98 - 1 ----------------GG-GA--CUG--GAGCUGCAAGUAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCUGAAAUUUUUGC-GC ----------------.(-..--(((--.((((((..........((.((((....)))).))....((((((....)))))))).))).).)))..).....((..........))-.. ( -29.10) >DroGri_CAF1 101333 112 - 1 UGGG------AAACUGGGCAA--CUGUAUAGCUGGGAGUAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCUGAAAUUUUUGCUGC .((.------...)).(((((--((((((.((((((.........((.((((....)))).))))).((((((....))))))...)))))))))..((......))......))))).. ( -34.30) >DroWil_CAF1 99785 104 - 1 ---UAG-------CC-AG-AG--UUG--GAGAUGCAAGUAGAUGACCCGCAUGAAAAUGCUGGCUCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCUGAAAUUUUUGCUGC ---(((-------((-(.-..--(((--....((((.(((((.(.((.((((....)))).))))))((((((....))))))....)).))))...))).))))))............. ( -35.00) >DroAna_CAF1 84603 100 - 1 -UGAAGCAA-------------------CGGCUCCAAGGAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCUGAAAUUUUUGCAGC -....((((-------------------((((((((.((.......))((((....))))))).(((((((((....))))))(((....))))))......)))))......))))... ( -28.80) >consensus __GGAG__________GG_AA__CUG__GAGCUGCAAGUAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCUGAAAUUUUUGCAGC .............................((((............((.((((....)))).)).(((((((((....))))))(((....))))))......)))).............. (-23.69 = -23.33 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:17 2006