| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,739,788 – 13,739,889 |
| Length | 101 |
| Max. P | 0.961794 |

| Location | 13,739,788 – 13,739,889 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 71.08 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -11.47 |
| Energy contribution | -11.22 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
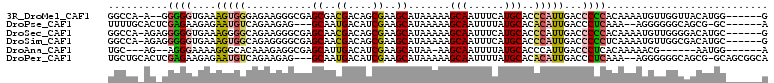
>3R_DroMel_CAF1 13739788 101 + 27905053 GGCCA-A--GGGGGUGAAAGUGGGAGAAGGGCGAGCGACGACAGCGAAGCAUAAAAAGCAAUUUCAUGCACCCAUUGACCCCCACAAAAUGUUGGUUACAUGG------G ..((.-.--((((((...((((((......((..((.......))...)).......(((......))).)))))).)))))).....((((.....))))))------. ( -32.00) >DroPse_CAF1 17034 98 + 1 UUUUGCACUCGAGAAGAGAAUGUCAGAAGAG---GCAAUGACAUCGAAGCAUAAAAAGCAAUUUUAUGCACACAUUGACCCUCAAA--AGGGGGGCAGCG-GC------A ...(((.(((.....))).((((((......---....))))))((..((((((((.....)))))))).....(((.(((((...--.))))).)))))-))------) ( -25.40) >DroSec_CAF1 13069 103 + 1 GGCCA-AGAGGGGGUGAAAGGGGCAGAAGGGCGAGCAACGACAGCGAAGCAUAAAAAGCAAUUUCAUGCACCCAUUGACCCCCACAAAAUGUUGGGGACAUGC------G .((..-...((((((.((.((((((((((.((..((..((....))..)).......))..)))).))).))).)).)))))).........((....)).))------. ( -32.90) >DroSim_CAF1 12756 103 + 1 GGCCA-AGAGGGGGUGAAAGUGGCAGAGGGGCGAGCAACGACAGCGAAGCAUAAAAAGCAAUUUCAUGCACCCAUUGACCCCCUCAAAAUGUUGGCGACAUGC------G .((((-(((((((((...(((((.......((..((.......))...)).......(((......)))..))))).))))))))......))))).......------. ( -33.20) >DroAna_CAF1 11563 92 + 1 UGC---AG--AGGGAAAAGGGCACAAAGAGGCGAGCAUUGACAUCGAAGCAUAA-AAGCAAUUUUAUGCACCCAUUGACCCUCACAAAAACG------AAUGG------A ...---.(--((((..(((((..........(((.........)))..((((((-((....)))))))).))).))..))))).........------.....------. ( -24.30) >DroPer_CAF1 16131 104 + 1 UGCUGCACUCGAGAAGAGAAUGUCAGAAGAG---GCAAUGACAUCGAAGCAUAAAAAGCAAUUUUAUGCACACAUUGACCCUCAAA--AGGGGGGCAGCG-GCAGCGGCA .(((((.(((.....))).((((((......---....))))))((..((((((((.....)))))))).....(((.(((((...--.))))).)))))-))))).... ( -32.90) >consensus GGCCA_AG_GGGGGAGAAAGUGGCAGAAGGGCGAGCAACGACAGCGAAGCAUAAAAAGCAAUUUCAUGCACCCAUUGACCCCCACAAAAUGGUGGCAACAUGC______A ..........((((....(((((...........((..((....))..)).......(((......)))..)))))...))))........................... (-11.47 = -11.22 + -0.25)
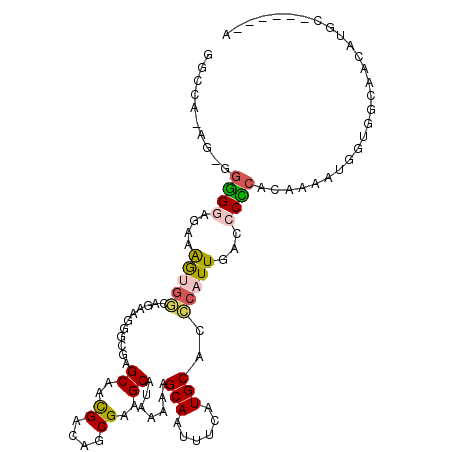
| Location | 13,739,788 – 13,739,889 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.08 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -13.07 |
| Energy contribution | -12.93 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13739788 101 - 27905053 C------CCAUGUAACCAACAUUUUGUGGGGGUCAAUGGGUGCAUGAAAUUGCUUUUUAUGCUUCGCUGUCGUCGCUCGCCCUUCUCCCACUUUCACCCCC--U-UGGCC .------((((((.....)))....(((((((.....((((((((((((.....))))))))...((.......))..))))..)))))))..........--.-))).. ( -27.60) >DroPse_CAF1 17034 98 - 1 U------GC-CGCUGCCCCCCU--UUUGAGGGUCAAUGUGUGCAUAAAAUUGCUUUUUAUGCUUCGAUGUCAUUGC---CUCUUCUGACAUUCUCUUCUCGAGUGCAAAA (------((-(((.....((((--....)))).....))).((((((((.....))))))))...(((((((....---......)))))))(((.....))).)))... ( -21.00) >DroSec_CAF1 13069 103 - 1 C------GCAUGUCCCCAACAUUUUGUGGGGGUCAAUGGGUGCAUGAAAUUGCUUUUUAUGCUUCGCUGUCGUUGCUCGCCCUUCUGCCCCUUUCACCCCCUCU-UGGCC .------((((((.....))))...(.((((((.((.((((((((((((.....))))))))...((.......))..........))))..)).)))))).).-..)). ( -28.80) >DroSim_CAF1 12756 103 - 1 C------GCAUGUCGCCAACAUUUUGAGGGGGUCAAUGGGUGCAUGAAAUUGCUUUUUAUGCUUCGCUGUCGUUGCUCGCCCCUCUGCCACUUUCACCCCCUCU-UGGCC .------.......(((((......((((((((.((..(((((((((((.....))))))))...((.......))..........)))...)).)))))))))-)))). ( -33.80) >DroAna_CAF1 11563 92 - 1 U------CCAUU------CGUUUUUGUGAGGGUCAAUGGGUGCAUAAAAUUGCUU-UUAUGCUUCGAUGUCAAUGCUCGCCUCUUUGUGCCCUUUUCCCU--CU---GCA .------.....------......((((((((.....((((((((((((....))-))))))..(((.((....))))).........))))....))))--).---))) ( -23.40) >DroPer_CAF1 16131 104 - 1 UGCCGCUGC-CGCUGCCCCCCU--UUUGAGGGUCAAUGUGUGCAUAAAAUUGCUUUUUAUGCUUCGAUGUCAUUGC---CUCUUCUGACAUUCUCUUCUCGAGUGCAGCA ....(((((-(((.....((((--....)))).....))).((((((((.....))))))))...(((((((....---......)))))))(((.....))).))))). ( -28.20) >consensus C______GCAUGUCGCCAACAUUUUGUGAGGGUCAAUGGGUGCAUAAAAUUGCUUUUUAUGCUUCGAUGUCAUUGCUCGCCCUUCUGCCACUUUCACCCCC_CU_UGGCA ...........................(((((((((((...((((((((.....))))))))........)))))...)))))).......................... (-13.07 = -12.93 + -0.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:42 2006