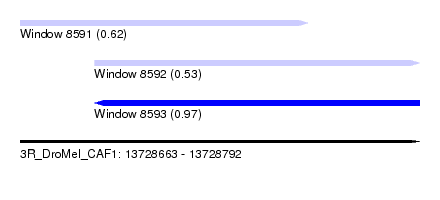
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,728,663 – 13,728,792 |
| Length | 129 |
| Max. P | 0.967216 |

| Location | 13,728,663 – 13,728,756 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -15.34 |
| Energy contribution | -16.23 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13728663 93 + 27905053 AAAUGACAGAAGGAGUUGAGUGGG-----------------------GGCGGUAAGCGGUGGGCGGUAUAAGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCA-GCAAUA ..............((((.(..((-----------------------..(((((((((.((.((.......)).))...)))))))).)...))..)((((....)))).-.)))). ( -28.30) >DroPse_CAF1 2545 87 + 1 AAAUGACAGCAGCUGGAGUGCG-------------------------GAAGGGGAGUGUUGGG-----GAAGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCAGGCAAUA ........((.((...((((((-------------------------.(((.((.((((((..-----......)))))).....)).....))).))))))...))....)).... ( -24.40) >DroSec_CAF1 1898 116 + 1 AAAUGACAGAAGGAGUUGUGUGGGCGGUGCGGGGAGAUGGCGGAUUGGGCGGUAAGCGGUGGGCGGUGUAAGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCA-GCAAUA ..............((((..(.((((.((((((......((.......))((((((((.((.((.......)).))...)))))))).....))))))))))...)..))-)).... ( -37.60) >DroEre_CAF1 1543 91 + 1 AAAUGACAGAAGGAGUUGUGUG-------------------------GGCGGUGGGCGGUGGGCGGUGUAUGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCA-GCAAUA ..............((((..(.-------------------------(((((..((.....(((((.(((((.....)))))))))).....))..).))))...)..))-)).... ( -29.50) >DroAna_CAF1 1854 86 + 1 AAAUGGUAGGGGAAUUUUUGGG---------------------------UGGU---UUCGGGGUGCUGCAAGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCA-GCAAUA ...(((((((((((((......---------------------------.)))---)))...(((.(((....))).))).))))))).....((((((((....)))).-)))).. ( -24.90) >DroPer_CAF1 2469 87 + 1 AAAUGACAGCAGCUGGAGAGGG-------------------------GGAGGGGAGUGUUGGG-----GAAGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCAGGCAAUA ........((.((.((.....(-------------------------(.(((...((((((..-----......)))))).))).)).....)).))((((....))))..)).... ( -22.20) >consensus AAAUGACAGAAGGAGUUGUGUG_________________________GGCGGUGAGCGGUGGGCGGUGUAAGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCA_GCAAUA ..................................................((((((((.((.((.......)).))...))))))))......((((((((....))))..)))).. (-15.34 = -16.23 + 0.89)


| Location | 13,728,687 – 13,728,792 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -24.37 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13728687 105 + 27905053 ----------GGCGGUAAGCGGUGGGCGGUAUAAGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCA-GCAAUAAAAAAUUUAAAUCACCAAGCGCCGCAUGUUGCACUG---- ----------(((((((((((.((.((.......)).))...))))))))......((((((((....)))).-))))......................)))((.....))....---- ( -32.90) >DroSec_CAF1 1938 112 + 1 CGGAU---UGGGCGGUAAGCGGUGGGCGGUGUAAGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCA-GCAAUAAAAAAUUUAAAUCACCAAGCGCCGCAUGUUGCACUG---- .((.(---(((..((((((((.((.((.......)).))...))))))))......((((((((....)))).-))))................))))...))((.....))....---- ( -34.70) >DroEre_CAF1 1565 105 + 1 ----------GGCGGUGGGCGGUGGGCGGUGUAUGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCA-GCAAUAAAAAAUUUAAAUCACCAAGCGCCGCAUGUUGCACUG---- ----------.((((((...(((((((((.(((((.....))))))))))......((((((((....)))).-))))..............))))...))))))...........---- ( -37.60) >DroYak_CAF1 1684 115 + 1 GUAAGCGGUGGGUGGUAGGCGGUGGGCGGUGUAAGUGCAACAUAGUUGCCAGAAACUUGCGCGCUAAUGUGUA-GCAAUAAAAAAUUUAAAUCACCAAGCGCCGCAUGUUGCACUG---- ((((((((((((((((..((.....)).(((((((((((((...))))).....))))))))((((.....))-))..............))))))...))))))...))))....---- ( -35.50) >DroAna_CAF1 1876 100 + 1 ------------UGGU---UUCGGGGUGCUGCAAGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCA-GCAAUAAAAAAUUGGAAUCACCCAGAACGGCAUGUUGCACAG---- ------------.(((---........)))....((((((((((((..........((((((((....)))).-)))).......((((......))))...))))))))))))..---- ( -32.70) >DroPer_CAF1 2491 105 + 1 ----------GGAGGGGAGUGUUGGG-----GAAGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCAGGCAAUAAAAAAUUUAAAUCACCAAGCGCCGCAUGUUGCAUACUUGU ----------.............(((-----...(((((((((((............((((((....))))))(((........................)))))))))))))).))).. ( -26.86) >consensus __________GGCGGUAAGCGGUGGGCGGUGUAAGUGCAACAUGCUUGCCAGAAACUUGCGCGCUAAUGUGCA_GCAAUAAAAAAUUUAAAUCACCAAGCGCCGCAUGUUGCACUG____ ..................................(((((((((((..((.((...)).))(((((...(((.....(((.....))).....)))..))))).)))))))))))...... (-24.37 = -24.90 + 0.53)



| Location | 13,728,687 – 13,728,792 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -29.59 |
| Energy contribution | -29.82 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
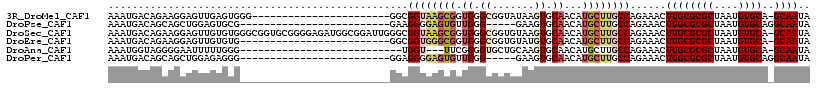
>3R_DroMel_CAF1 13728687 105 - 27905053 ----CAGUGCAACAUGCGGCGCUUGGUGAUUUAAAUUUUUUAUUGC-UGCACAUUAGCGCGCAAGUUUCUGGCAAGCAUGUUGCACUUAUACCGCCCACCGCUUACCGCC---------- ----.((((((((((((.(((((..(((.....(((.....)))..-..)))...)))))((.((...)).))..)))))))))))).............((.....)).---------- ( -33.80) >DroSec_CAF1 1938 112 - 1 ----CAGUGCAACAUGCGGCGCUUGGUGAUUUAAAUUUUUUAUUGC-UGCACAUUAGCGCGCAAGUUUCUGGCAAGCAUGUUGCACUUACACCGCCCACCGCUUACCGCCCA---AUCCG ----.((((((((((((.(((((..(((.....(((.....)))..-..)))...)))))((.((...)).))..)))))))))))).............((.....))...---..... ( -33.80) >DroEre_CAF1 1565 105 - 1 ----CAGUGCAACAUGCGGCGCUUGGUGAUUUAAAUUUUUUAUUGC-UGCACAUUAGCGCGCAAGUUUCUGGCAAGCAUGUUGCACAUACACCGCCCACCGCCCACCGCC---------- ----..(((((((((((.(((((..(((.....(((.....)))..-..)))...)))))((.((...)).))..)))))))))))........................---------- ( -33.40) >DroYak_CAF1 1684 115 - 1 ----CAGUGCAACAUGCGGCGCUUGGUGAUUUAAAUUUUUUAUUGC-UACACAUUAGCGCGCAAGUUUCUGGCAACUAUGUUGCACUUACACCGCCCACCGCCUACCACCCACCGCUUAC ----.(((((((((((((.((((..(((.....(((.....)))..-..)))...)))))))........(....).))))))))))................................. ( -27.20) >DroAna_CAF1 1876 100 - 1 ----CUGUGCAACAUGCCGUUCUGGGUGAUUCCAAUUUUUUAUUGC-UGCACAUUAGCGCGCAAGUUUCUGGCAAGCAUGUUGCACUUGCAGCACCCCGAA---ACCA------------ ----..(((((((((((.....((((....))))........((((-(((......))((....))....))))))))))))))))...............---....------------ ( -30.10) >DroPer_CAF1 2491 105 - 1 ACAAGUAUGCAACAUGCGGCGCUUGGUGAUUUAAAUUUUUUAUUGCCUGCACAUUAGCGCGCAAGUUUCUGGCAAGCAUGUUGCACUUC-----CCCAACACUCCCCUCC---------- .......((((((((((.(((((..(((.....(((.....))).....)))...)))))((.((...)).))..))))))))))....-----................---------- ( -30.00) >consensus ____CAGUGCAACAUGCGGCGCUUGGUGAUUUAAAUUUUUUAUUGC_UGCACAUUAGCGCGCAAGUUUCUGGCAAGCAUGUUGCACUUACACCGCCCACCGCUUACCGCC__________ ......(((((((((((.(((((..(((.....(((.....))).....)))...)))))((.((...)).))..))))))))))).................................. (-29.59 = -29.82 + 0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:37 2006